
| Microexon ID | Gm_1:399995-400005:- |
| Species | Glycine max | Coordinates | 1:399995..400005 |
| Microexon Cluster ID | MEP26 |
| Size | 11 |
| Phase | 1 |
| Pfam Domain Motif | PRMT5 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,11,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RRYGAYAARACCAGYGCYGAYTAYTACTTCGAYTCCTACTCYCAYTTYGGTATTCATGAAGAAATGYTGAARGATRYWGTGAGRACWAARACWTAYCAAAATGTTATY |
| Logo of Microexon-tag DNA Seq | 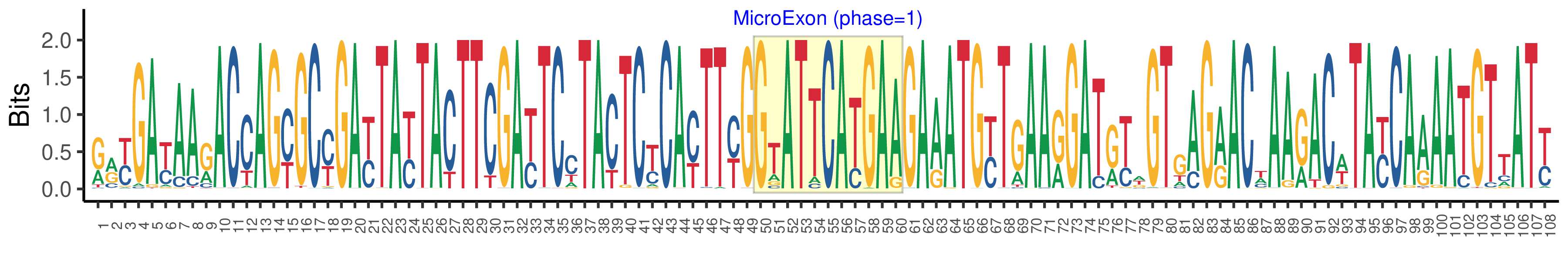 |
| Alignment of exons | 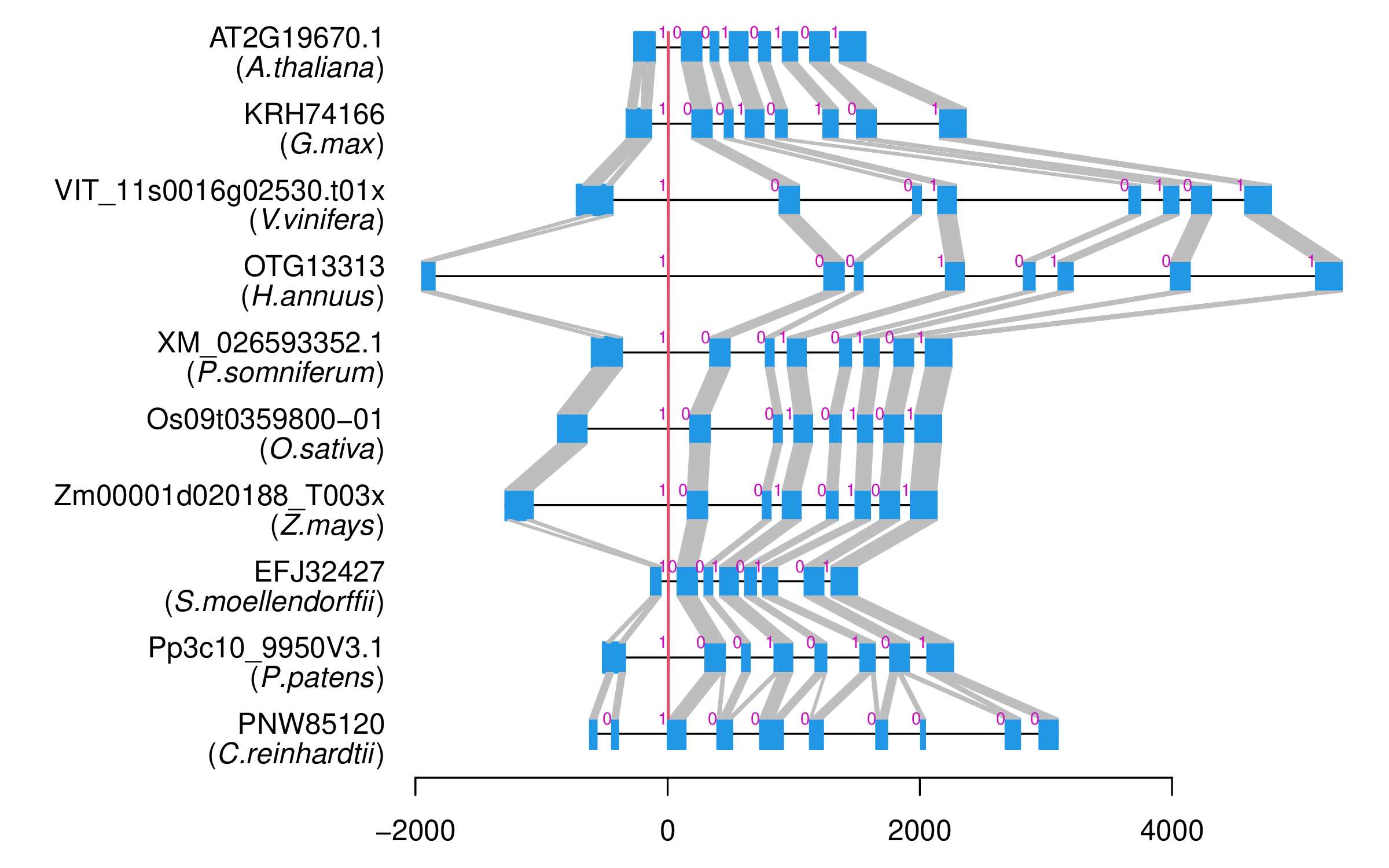 |
| Microexon DNA seq | GAATTCACGAA |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | GATGATAAAACCAGCGCCGACTATTACTTCGATTCCTACTCTCATTTTGGAATTCACGAAGAGATGTTGAAGGACACTGTGAGAACCAAGACATATCAAAATGTTATT |
| Microexon-tag Amino Acid Seq | DDKTSADYYFDSYSHFGIHEEMLKDTVRTKTYQNVI |
| Microexon-tag spanning region | 399763-400183 |
| Microexon-tag prediction score | 0.9817 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH74166x |
| Reference Transcript ID | KRH74166 |
| Gene ID | GLYMA_01G003800 |
| Gene Name | NA |
| Transcript ID | KRH74166 |
| Protein ID | KRH74166 |
| Gene ID | GLYMA_01G003800 |
| Gene Name | NA |
| Pfam domain motif | PRMT5 |
| Motif E-value | 0.00013 |
| Motif start | 69 |
| Motif end | 172 |
| Protein seq | >KRH74166 MGQRKNDNNINQCSSSKEDADMNNNHLRFEEAVDESSNLDQSMCDIEESDDKTSADYYFDSYSHFGIHEEMLKDTVRTKT YQNVIYQNKFLFKNKVVLDVGAGTGILSLFCAKAGAEHVYAVECSHMADMAKEIVEANGYSNVVTVLKGKIEEIELPVAK VDIIISEWMGYFLLFENMLNSVLYARDKWLVDGGVVLPDKASLHLTAIEDADYKEDKIEFWNNVYGFDMSCIKKQAIMEP LVDTVDQNQIATNCQLLKTMDISKMAPGDASFTVPFKLVAERDDYIHALVAYFDVSFTKCHKLMGFSTGPRSRATHWKQT VLYLEDVLTICEGEAIVGSMAVAPNKKNPRDVDIMLKYSLNGRRCNVSRVQYYKMR* |
| CDS seq | >KRH74166 ATGGGTCAGCGAAAGAACGACAACAACATCAATCAGTGCTCCTCAAGCAAGGAAGACGCTGACATGAATAACAACCACCT TCGCTTTGAAGAGGCTGTCGACGAGAGTTCCAACCTCGACCAATCCATGTGCGACATCGAAGAATCCGATGATAAAACCA GCGCCGACTATTACTTCGATTCCTACTCTCATTTTGGAATTCACGAAGAGATGTTGAAGGACACTGTGAGAACCAAGACA TATCAAAATGTTATTTATCAGAACAAGTTTTTATTCAAGAATAAAGTAGTTCTTGATGTTGGTGCTGGGACTGGGATTTT ATCACTATTTTGTGCCAAAGCAGGGGCAGAACACGTCTATGCGGTTGAGTGCTCCCACATGGCTGACATGGCAAAGGAGA TTGTTGAAGCTAATGGTTACTCTAATGTTGTAACAGTTTTGAAGGGGAAGATTGAAGAAATTGAACTTCCAGTTGCTAAA GTTGATATAATTATTTCAGAATGGATGGGATATTTCTTGTTGTTTGAGAATATGTTAAATTCGGTGCTCTATGCTCGTGA CAAATGGCTTGTGGATGGCGGAGTTGTGCTACCAGACAAAGCATCCCTCCATCTCACTGCTATTGAAGATGCTGACTATA AAGAAGATAAAATTGAGTTTTGGAACAATGTATATGGATTTGACATGAGCTGCATCAAGAAGCAAGCCATAATGGAGCCT CTTGTTGACACAGTCGACCAGAATCAGATTGCTACAAACTGTCAGCTACTCAAGACAATGGATATCTCAAAGATGGCTCC TGGAGATGCTTCATTCACAGTACCTTTTAAGCTTGTAGCTGAACGTGATGACTATATTCATGCTCTTGTTGCATATTTTG ATGTATCGTTTACAAAGTGTCATAAATTGATGGGCTTCTCTACAGGACCAAGATCACGAGCTACGCATTGGAAACAAACA GTCCTATACCTGGAAGATGTCTTGACCATTTGTGAGGGGGAGGCAATTGTGGGGAGCATGGCTGTTGCTCCAAATAAAAA AAATCCTCGGGATGTTGATATAATGCTCAAGTATTCATTGAATGGAAGGCGATGCAATGTTTCAAGGGTTCAGTACTACA AGATGCGTTGA |
| Microexon DNA seq | GAATTCACGAA |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | GATGATAAAACCAGCGCCGACTATTACTTCGATTCCTACTCTCATTTTGGAATTCACGAAGAGATGTTGAAGGACACTGTGAGAACCAAGACATATCAAAATGTTATT |
| Microexon-tag Amino Acid seq | DDKTSADYYFDSYSHFGIHEEMLKDTVRTKTYQNVI |
| Transcript ID | KRH74166 |
| Gene ID | Gm.44 |
| Gene Name | NA |
| Pfam domain motif | PRMT5 |
| Motif E-value | 0.00013 |
| Motif start | 69 |
| Motif end | 172 |
| Protein seq | >KRH74166 MGQRKNDNNINQCSSSKEDADMNNNHLRFEEAVDESSNLDQSMCDIEESDDKTSADYYFDSYSHFGIHEEMLKDTVRTKT YQNVIYQNKFLFKNKVVLDVGAGTGILSLFCAKAGAEHVYAVECSHMADMAKEIVEANGYSNVVTVLKGKIEEIELPVAK VDIIISEWMGYFLLFENMLNSVLYARDKWLVDGGVVLPDKASLHLTAIEDADYKEDKIEFWNNVYGFDMSCIKKQAIMEP LVDTVDQNQIATNCQLLKTMDISKMAPGDASFTVPFKLVAERDDYIHALVAYFDVSFTKCHKLMGFSTGPRSRATHWKQT VLYLEDVLTICEGEAIVGSMAVAPNKKNPRDVDIMLKYSLNGRRCNVSRVQYYKMR* |
| CDS seq | >KRH74166 ATGGGTCAGCGAAAGAACGACAACAACATCAATCAGTGCTCCTCAAGCAAGGAAGACGCTGACATGAATAACAACCACCT TCGCTTTGAAGAGGCTGTCGACGAGAGTTCCAACCTCGACCAATCCATGTGCGACATCGAAGAATCCGATGATAAAACCA GCGCCGACTATTACTTCGATTCCTACTCTCATTTTGGAATTCACGAAGAGATGTTGAAGGACACTGTGAGAACCAAGACA TATCAAAATGTTATTTATCAGAACAAGTTTTTATTCAAGAATAAAGTAGTTCTTGATGTTGGTGCTGGGACTGGGATTTT ATCACTATTTTGTGCCAAAGCAGGGGCAGAACACGTCTATGCGGTTGAGTGCTCCCACATGGCTGACATGGCAAAGGAGA TTGTTGAAGCTAATGGTTACTCTAATGTTGTAACAGTTTTGAAGGGGAAGATTGAAGAAATTGAACTTCCAGTTGCTAAA GTTGATATAATTATTTCAGAATGGATGGGATATTTCTTGTTGTTTGAGAATATGTTAAATTCGGTGCTCTATGCTCGTGA CAAATGGCTTGTGGATGGCGGAGTTGTGCTACCAGACAAAGCATCCCTCCATCTCACTGCTATTGAAGATGCTGACTATA AAGAAGATAAAATTGAGTTTTGGAACAATGTATATGGATTTGACATGAGCTGCATCAAGAAGCAAGCCATAATGGAGCCT CTTGTTGACACAGTCGACCAGAATCAGATTGCTACAAACTGTCAGCTACTCAAGACAATGGATATCTCAAAGATGGCTCC TGGAGATGCTTCATTCACAGTACCTTTTAAGCTTGTAGCTGAACGTGATGACTATATTCATGCTCTTGTTGCATATTTTG ATGTATCGTTTACAAAGTGTCATAAATTGATGGGCTTCTCTACAGGACCAAGATCACGAGCTACGCATTGGAAACAAACA GTCCTATACCTGGAAGATGTCTTGACCATTTGTGAGGGGGAGGCAATTGTGGGGAGCATGGCTGTTGCTCCAAATAAAAA AAATCCTCGGGATGTTGATATAATGCTCAAGTATTCATTGAATGGAAGGCGATGCAATGTTTCAAGGGTTCAGTACTACA AGATGCGTTGA |