
| Microexon ID | Bn_scaffoldC07:4263238-4263248:+ |
| Species | Brassica Napus | Coordinates | scaffoldC07:4263238..4263248 |
| Microexon Cluster ID | MEP26 |
| Size | 11 |
| Phase | 1 |
| Pfam Domain Motif | PRMT5 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,11,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RRYGAYAARACCAGYGCYGAYTAYTACTTCGAYTCCTACTCYCAYTTYGGTATTCATGAAGAAATGYTGAARGATRYWGTGAGRACWAARACWTAYCAAAATGTTATY |
| Logo of Microexon-tag DNA Seq | 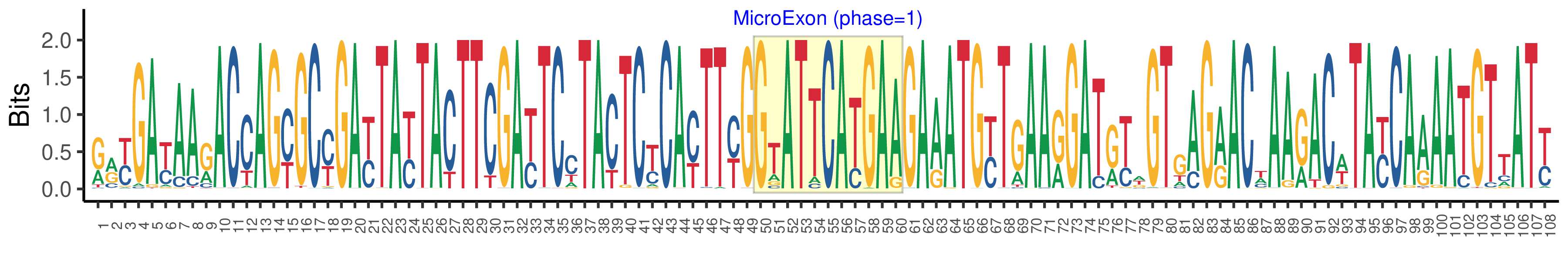 |
| Alignment of exons |  |
| Microexon DNA seq | GTATTCATGAA |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | GATGACATAACCAGCGCCGATTACTACTTCGATTCTTACTCCCATTTCGGTATTCATGAAGAAATGTTGAAGGATGTAGTGAGGACAAAGAGTTACCGTGATGTTATT |
| Microexon-tag Amino Acid Seq | DDITSADYYFDSYSHFGIHEEMLKDVVRTKSYRDVI |
| Microexon-tag spanning region | 4263089-4263371 |
| Microexon-tag prediction score | 0.9633 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | BnaC07T0023100ZSx |
| Reference Transcript ID | BnaC07T0023100ZS |
| Gene ID | BnaC07G0023100ZS |
| Gene Name | NA |
| Transcript ID | BnaC07T0023100ZS |
| Protein ID | |
| Gene ID | BnaC07G0023100ZS |
| Gene Name | |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >BnaC07T0023100ZS MAATKNNHNELGSDQHTKLPFEDVDETMCDADVADDITSADYYFDSYSHFGIHEEMLKDVVRTKSYRDVIYKNKFLVKDK IVLDVGAGTGILSLFCAKAGAAHVYAVECSQMADTAKEIVKSNGFSDGNNLIQMCKVFINNFVTNNCLVIFGVVITVLKG KIEEIELPVPKVDVIISEWMGYFLVYENMLDTVLYARNKWLVDGGIVLPDKASLYLTAIEDAHYKEDKVEFWNDVYGFDM SCIKRRAITEPLVDTVDGNQIVTDSKLLKTMDISKMASEDASFTAPFKLVAKRNDHIHALVAYFDVSFTMCHKMIGFSTG PKSRATHWKQTVLYLEDVLTICEGEMITGSMTIAPNKKNPRDVDIELSYSLNGQHCKISRTQLYKMR* |
| CDS seq | >BnaC07T0023100ZS ATGGCTGCTACGAAGAACAACCACAACGAGCTAGGCTCTGATCAACACACGAAGTTACCCTTTGAAGACGTCGATGAAAC AATGTGCGATGCCGATGTAGCTGATGACATAACCAGCGCCGATTACTACTTCGATTCTTACTCCCATTTCGGTATTCATG AAGAAATGTTGAAGGATGTAGTGAGGACAAAGAGTTACCGTGATGTTATTTACAAGAACAAGTTTCTTGTCAAGGACAAA ATCGTTCTTGATGTTGGAGCTGGGACAGGGATCTTGTCTCTCTTCTGTGCTAAAGCAGGAGCTGCTCACGTATACGCTGT TGAATGTTCTCAAATGGCTGACACTGCGAAGGAGATCGTCAAGTCGAATGGGTTTTCTGATGGTAATAATCTGATACAAA TGTGTAAAGTGTTCATAAATAACTTTGTTACTAACAACTGTTTGGTTATTTTTGGGGTAGTTATAACGGTTTTGAAAGGG AAGATTGAGGAAATCGAGCTTCCAGTTCCTAAAGTGGATGTGATTATCTCTGAGTGGATGGGTTACTTTCTTGTTTATGA AAACATGCTTGACACCGTCTTGTATGCTCGCAATAAATGGCTTGTCGATGGTGGAATTGTTCTACCAGATAAAGCTTCTC TCTATCTTACAGCCATAGAGGATGCTCATTACAAAGAAGACAAAGTTGAATTTTGGAACGACGTGTATGGGTTTGACATG TCATGCATCAAGAGAAGAGCAATCACGGAGCCTCTTGTTGATACAGTTGATGGCAACCAAATCGTAACTGATAGCAAGCT ACTCAAGACGATGGATATCTCTAAGATGGCTTCTGAAGACGCTTCCTTTACAGCTCCGTTCAAGCTTGTGGCAAAACGGA ATGATCATATTCATGCACTTGTAGCCTACTTTGATGTATCATTCACTATGTGCCACAAGATGATTGGTTTCTCAACAGGA CCAAAATCTAGGGCTACACACTGGAAGCAAACAGTGCTATACCTTGAAGATGTGTTAACCATATGCGAGGGTGAAATGAT CACAGGAAGCATGACTATTGCACCTAACAAGAAGAATCCTAGAGATGTGGACATAGAGCTCAGTTATTCTTTGAATGGCC AGCATTGCAAGATCTCAAGGACCCAACTCTACAAAATGCGGTAA |
Bn_scaffoldC07:4263238-4263248:+ does not have available information here.