
| Microexon ID | Bn_scaffoldA08:20232039-20232049:- |
| Species | Brassica Napus | Coordinates | scaffoldA08:20232039..20232049 |
| Microexon Cluster ID | MEP26 |
| Size | 11 |
| Phase | 1 |
| Pfam Domain Motif | PRMT5 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,11,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RRYGAYAARACCAGYGCYGAYTAYTACTTCGAYTCCTACTCYCAYTTYGGTATTCATGAAGAAATGYTGAARGATRYWGTGAGRACWAARACWTAYCAAAATGTTATY |
| Logo of Microexon-tag DNA Seq | 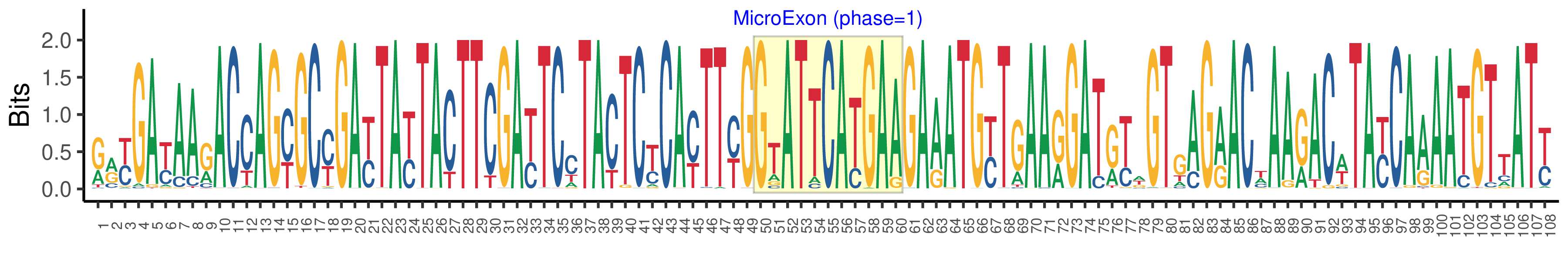 |
| Alignment of exons | 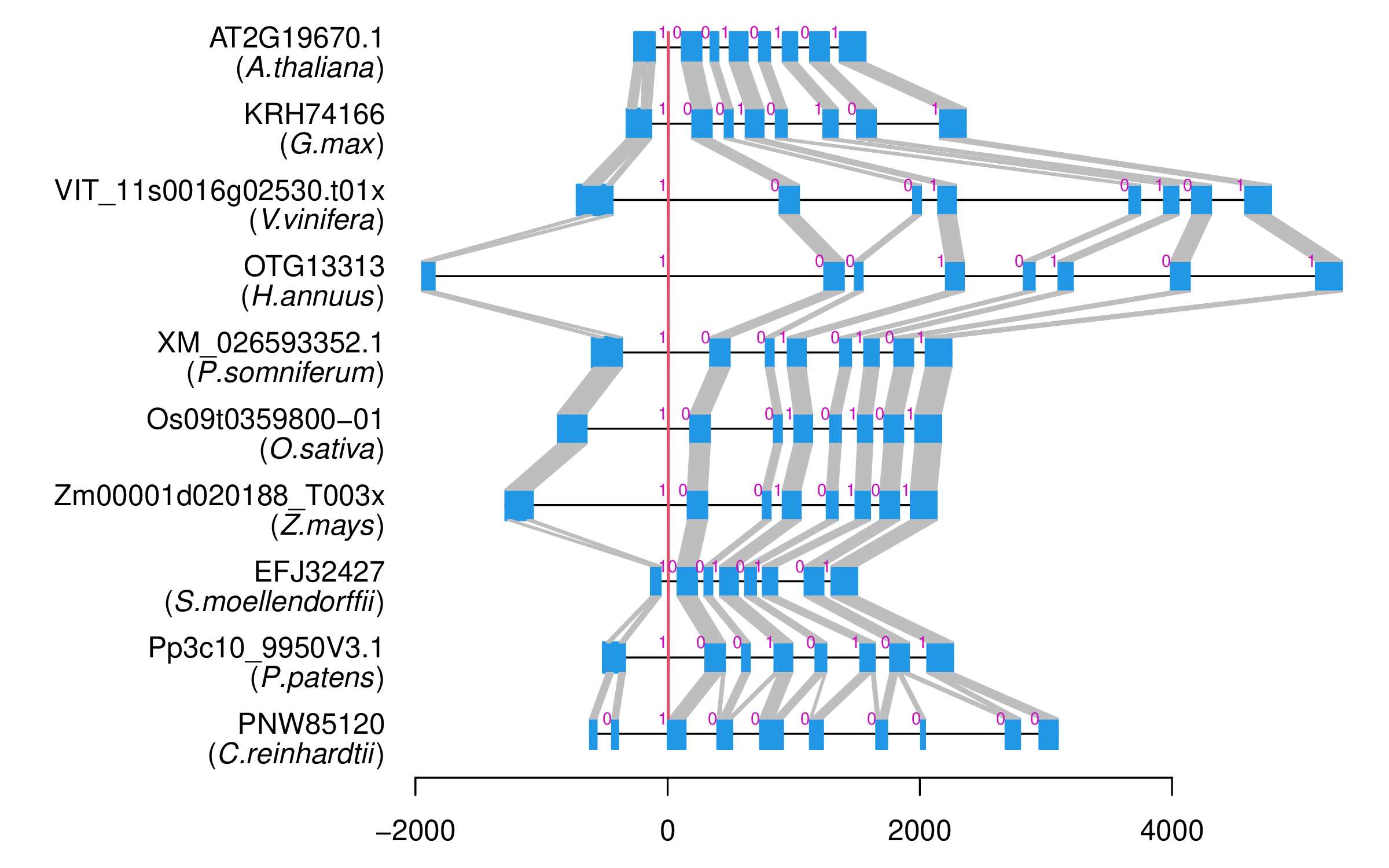 |
| Microexon DNA seq | GAATTCACGAA |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | GATGATACCACCAGCGCCGACTACTACTTCGATTCCTACTCTCACTTCGGAATTCACGAAGAGATGTTGAAGGATGTAGTGAGAACAAAGACTTACCAGAATGTAATT |
| Microexon-tag Amino Acid Seq | DDTTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNVI |
| Microexon-tag spanning region | 20231896-20232194 |
| Microexon-tag prediction score | 0.9845 |
| Overlapped with the annotated transcript (%) | 94.44 |
| New Transcript ID | BnaA08T0159000ZSx |
| Reference Transcript ID | BnaA08T0159000ZS |
| Gene ID | BnaA08G0159000ZS |
| Gene Name | NA |
| Transcript ID | BnaA08T0159000ZS |
| Protein ID | |
| Gene ID | BnaA08G0159000ZS |
| Gene Name | |
| Pfam domain motif | PRMT5 |
| Motif E-value | 1.00E-05 |
| Motif start | 40 |
| Motif end | 135 |
| Protein seq | >BnaA08T0159000ZS MKSNNEHQVAQGAQDESMCDVTDDTTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNDKVVLDVGAGTGILSLFCAKAGAA HVYAVECSQMADMAKEIVKANGFSDVITVLKGKIEEIELPTPKVDVIISEWMGYFLLFENMLDSVL* |
| CDS seq | >BnaA08T0159000ZS ATGAAGAGTAACAACGAACACCAAGTTGCACAAGGCGCCCAAGATGAATCAATGTGCGACGTCACTGATGATACCACCAG CGCCGACTACTACTTCGATTCCTACTCTCACTTCGGAATTCACGAAGAGATGTTGAAGGATGTAGTGAGAACAAAGACTT ACCAGAATGACAAAGTTGTTCTTGATGTTGGAGCTGGGACTGGAATCTTGTCTCTCTTCTGTGCCAAAGCAGGAGCTGCT CATGTCTACGCTGTTGAATGTTCTCAAATGGCTGACATGGCAAAGGAGATTGTTAAAGCCAACGGCTTCTCTGATGTGAT AACGGTATTGAAAGGGAAGATTGAGGAGATAGAGCTTCCTACTCCTAAAGTGGATGTGATTATATCTGAATGGATGGGTT ACTTTCTGTTGTTTGAGAATATGTTGGACAGTGTCTTATAA |
Bn_scaffoldA08:20232039-20232049:- does not have available information here.