
| Microexon ID | Zm_5:72779203-72779212:+ |
| Species | Zea mays | Coordinates | 5:72779203..72779212 |
| Microexon Cluster ID | MEP25 |
| Size | 10 |
| Phase | 2 |
| Pfam Domain Motif | CDP-OH_P_transf |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 50,10,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | YTSCARCCYTTYTGGASYCGHTKYGTYAMYYTCTTCCCYCTTTGGATGCCRCCAAAYATGATWACACTTAYRGGATTYATGTTYYTRSTBAYATCTGCAYTGCTTGGC |
| Logo of Microexon-tag DNA Seq | 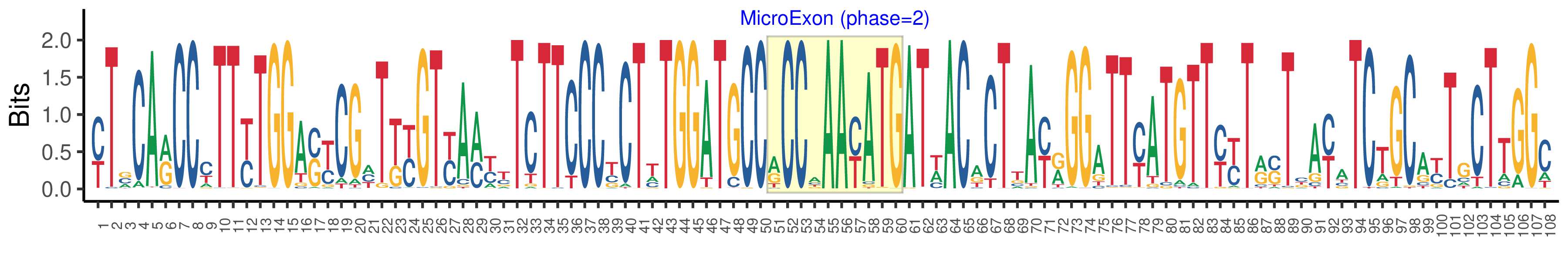 |
| Alignment of exons | 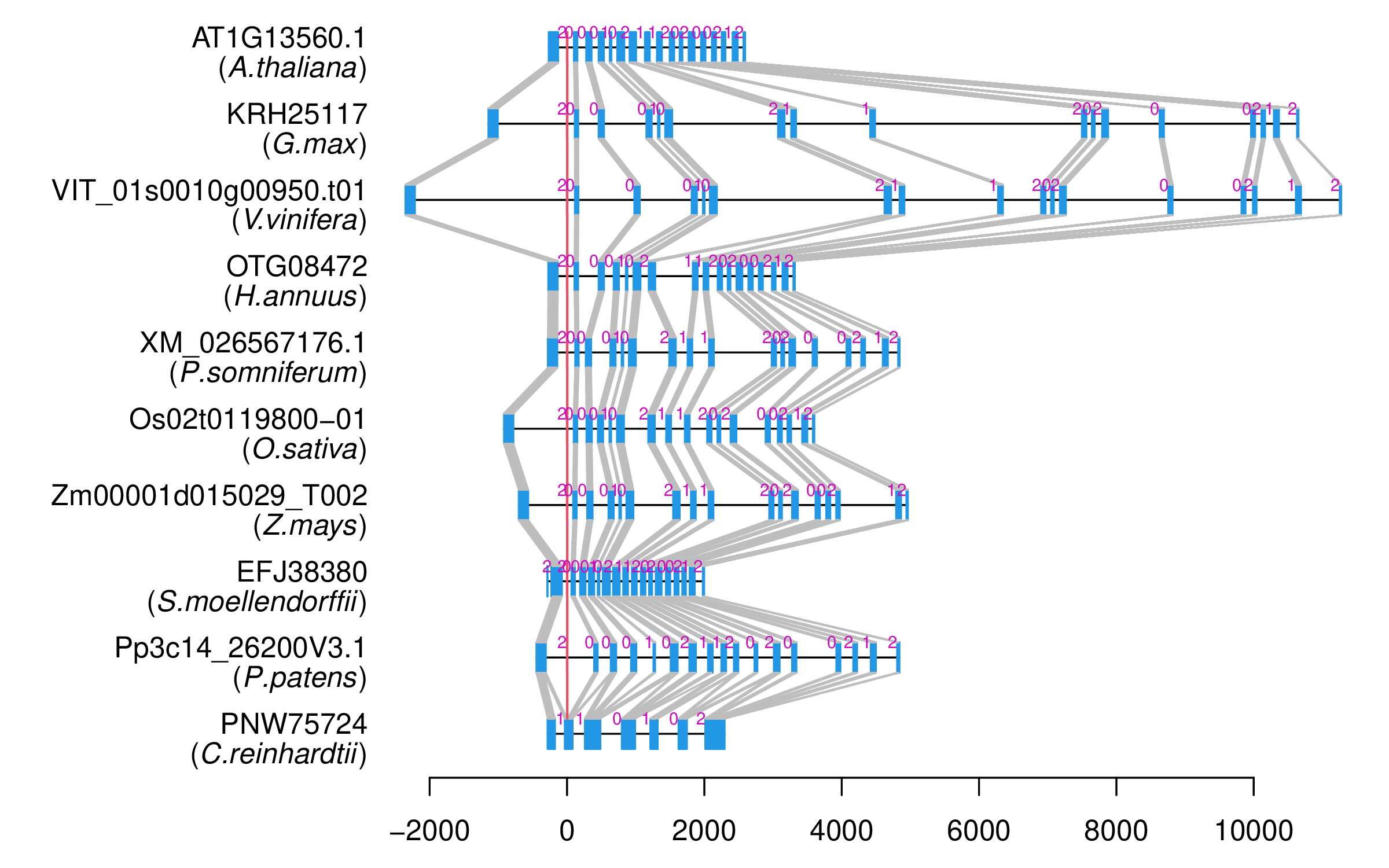 |
| Microexon DNA seq | ACCGAACATG |
| Microexon Amino Acid seq | PPNM |
| Microexon-tag DNA Seq | CTCCAGCCCTTCTGGTCCCGATTCGTCAACATCTTCCCGCTATGGTTCCCACCGAACATGATTACACTGACAGGTTTCATGTTTCTACTGACATCAGCATTCCTTGGC |
| Microexon-tag Amino Acid Seq | LQPFWSRFVNIFPLWFPPNMITLTGFMFLLTSAFLG |
| Microexon-tag spanning region | 72778590-72779342 |
| Microexon-tag prediction score | 0.9682 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Zm00001d015029_T002x |
| Reference Transcript ID | Zm00001d015029_T002 |
| Gene ID | Zm00001d015029 |
| Gene Name | NA |
| Transcript ID | Zm00001d015029_T002 |
| Protein ID | Zm00001d015029_P002 |
| Gene ID | Zm00001d015029 |
| Gene Name | NA |
| Pfam domain motif | CDP-OH_P_transf |
| Motif E-value | 1.7e-16 |
| Motif start | 47 |
| Motif end | 120 |
| Protein seq | >Zm00001d015029_P002 MGYIGHHGVATLRRYKYSGVDHSLVAKYILQPFWSRFVNIFPLWFPPNMITLTGFMFLLTSAFLGFLYSPHLDTAPPRWV HLAHGLLLFLYQTFDAVDGKQARRTNSSSPLGELFDHGCDALACAFESLAFGSTAMCGKATFWFWFISAVPFYFATWEHF FTNTLILPIVNGPTEGLMLIYLCHFFTFFTGAEWWAQDFQKSMPLLGWVPLISEIPVYDIVLCLMIAFAVIPTIGSNIHN VYKVVEARKGSMLLALAMLFPFGLLLAGVLVWSYLSPSDIMRTQPHLLIIGTGFAFGFLVGRMILAHLCDEPKGLKTGMC MSLAYFPFAIANALTARLDDGNPLVDEQLVLLMYCLFTVALYMHFATSVIHEITNALGIHCFRITRKKA* |
| CDS seq | >Zm00001d015029_T002 ATGGGTTACATTGGCCACCACGGCGTCGCCACGCTGCGGCGCTACAAGTACAGCGGCGTCGACCACTCGCTCGTCGCCAA GTACATCCTCCAGCCCTTCTGGTCCCGATTCGTCAACATCTTCCCGCTATGGTTCCCACCGAACATGATTACACTGACAG GTTTCATGTTTCTACTGACATCAGCATTCCTTGGCTTTTTATATTCACCCCATCTAGATACAGCACCCCCCAGATGGGTT CACCTTGCTCATGGACTGCTTCTCTTTCTTTATCAGACTTTTGATGCTGTGGATGGAAAACAAGCAAGGCGTACCAACTC ATCAAGTCCTCTAGGCGAGCTTTTTGACCACGGATGTGATGCGCTTGCGTGTGCTTTTGAATCCTTGGCTTTTGGAAGCA CGGCGATGTGTGGAAAGGCTACCTTCTGGTTTTGGTTCATTTCAGCTGTCCCATTTTACTTCGCAACCTGGGAACACTTT TTTACAAATACACTTATTCTTCCTATAGTCAACGGACCAACTGAAGGCCTTATGCTGATCTACTTGTGCCATTTTTTCAC CTTTTTCACAGGAGCCGAGTGGTGGGCACAGGATTTTCAGAAGTCAATGCCCCTGCTGGGTTGGGTTCCTCTTATCTCTG AAATCCCGGTGTATGACATTGTGCTATGTCTTATGATTGCTTTTGCTGTAATCCCAACAATTGGATCTAACATCCACAAT GTATATAAAGTCGTTGAAGCAAGAAAAGGAAGCATGCTTCTGGCACTAGCCATGCTCTTTCCTTTTGGTTTGCTCTTGGC TGGAGTTCTTGTCTGGTCCTACCTTTCTCCTTCAGATATAATGAGAACCCAACCACATTTGCTAATTATTGGAACTGGTT TTGCATTTGGATTTCTTGTGGGAAGAATGATTCTGGCTCACTTGTGTGATGAACCCAAGGGTTTGAAAACAGGGATGTGC ATGTCCCTTGCGTATTTTCCATTTGCAATTGCAAACGCGTTGACTGCCCGGCTTGATGATGGAAATCCACTCGTTGATGA GCAGCTAGTGCTCCTGATGTACTGCCTATTTACAGTGGCTCTGTACATGCATTTTGCTACATCAGTTATTCATGAGATCA CCAATGCACTCGGGATCCACTGCTTCAGGATCACTAGGAAAAAGGCATAG |
| Microexon DNA seq | ACCGAACATG |
| Microexon Amino Acid seq | PPNM |
| Microexon-tag DNA Seq | CTCCAGCCCTTCTGGTCCCGATTCGTCAACATCTTCCCGCTATGGTTCCCACCGAACATGATTACACTGACAGGTTTCATGTTTCTACTGACATCAGCATTCCTTGGC |
| Microexon-tag Amino Acid seq | LQPFWSRFVNIFPLWFPPNMITLTGFMFLLTSAFLG |
| Transcript ID | Zm00001d015029_T002 |
| Gene ID | Zm.23315 |
| Gene Name | NA |
| Pfam domain motif | CDP-OH_P_transf |
| Motif E-value | 1.7e-16 |
| Motif start | 47 |
| Motif end | 120 |
| Protein seq | >Zm00001d015029_T002 MGYIGHHGVATLRRYKYSGVDHSLVAKYILQPFWSRFVNIFPLWFPPNMITLTGFMFLLTSAFLGFLYSPHLDTAPPRWV HLAHGLLLFLYQTFDAVDGKQARRTNSSSPLGELFDHGCDALACAFESLAFGSTAMCGKATFWFWFISAVPFYFATWEHF FTNTLILPIVNGPTEGLMLIYLCHFFTFFTGAEWWAQDFQKSMPLLGWVPLISEIPVYDIVLCLMIAFAVIPTIGSNIHN VYKVVEARKGSMLLALAMLFPFGLLLAGVLVWSYLSPSDIMRTQPHLLIIGTGFAFGFLVGRMILAHLCDEPKGLKTGMC MSLAYFPFAIANALTARLDDGNPLVDEQLVLLMYCLFTVALYMHFATSVIHEITNALGIHCFRITRKKA* |
| CDS seq | >Zm00001d015029_T002 ATGGGTTACATTGGCCACCACGGCGTCGCCACGCTGCGGCGCTACAAGTACAGCGGCGTCGACCACTCGCTCGTCGCCAA GTACATCCTCCAGCCCTTCTGGTCCCGATTCGTCAACATCTTCCCGCTATGGTTCCCACCGAACATGATTACACTGACAG GTTTCATGTTTCTACTGACATCAGCATTCCTTGGCTTTTTATATTCACCCCATCTAGATACAGCACCCCCCAGATGGGTT CACCTTGCTCATGGACTGCTTCTCTTTCTTTATCAGACTTTTGATGCTGTGGATGGAAAACAAGCAAGGCGTACCAACTC ATCAAGTCCTCTAGGCGAGCTTTTTGACCACGGATGTGATGCGCTTGCGTGTGCTTTTGAATCCTTGGCTTTTGGAAGCA CGGCGATGTGTGGAAAGGCTACCTTCTGGTTTTGGTTCATTTCAGCTGTCCCATTTTACTTCGCAACCTGGGAACACTTT TTTACAAATACACTTATTCTTCCTATAGTCAACGGACCAACTGAAGGCCTTATGCTGATCTACTTGTGCCATTTTTTCAC CTTTTTCACAGGAGCCGAGTGGTGGGCACAGGATTTTCAGAAGTCAATGCCCCTGCTGGGTTGGGTTCCTCTTATCTCTG AAATCCCGGTGTATGACATTGTGCTATGTCTTATGATTGCTTTTGCTGTAATCCCAACAATTGGATCTAACATCCACAAT GTATATAAAGTCGTTGAAGCAAGAAAAGGAAGCATGCTTCTGGCACTAGCCATGCTCTTTCCTTTTGGTTTGCTCTTGGC TGGAGTTCTTGTCTGGTCCTACCTTTCTCCTTCAGATATAATGAGAACCCAACCACATTTGCTAATTATTGGAACTGGTT TTGCATTTGGATTTCTTGTGGGAAGAATGATTCTGGCTCACTTGTGTGATGAACCCAAGGGTTTGAAAACAGGGATGTGC ATGTCCCTTGCGTATTTTCCATTTGCAATTGCAAACGCGTTGACTGCCCGGCTTGATGATGGAAATCCACTCGTTGATGA GCAGCTAGTGCTCCTGATGTACTGCCTATTTACAGTGGCTCTGTACATGCATTTTGCTACATCAGTTATTCATGAGATCA CCAATGCACTCGGGATCCACTGCTTCAGGATCACTAGGAAAAAGGCATAG |