
| Microexon ID | Sm_GL377565:4057862-4057871:+ |
| Species | Selaginella moellendorffii | Coordinates | GL377565:4057862..4057871 |
| Microexon Cluster ID | MEP24 |
| Size | 10 |
| Phase | 1 |
| Pfam Domain Motif | Ham1p_like |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,10,32,17 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | YTKGTKGARGAYACTTGYCTCTGYTTCAATGCMYTCAARGGTCTTCCAGGGCCYTACATMAARTGGTTTCTGSAGAAGATTGGTCATGAAGGTYTGAACAAYTTGYTR |
| Logo of Microexon-tag DNA Seq | 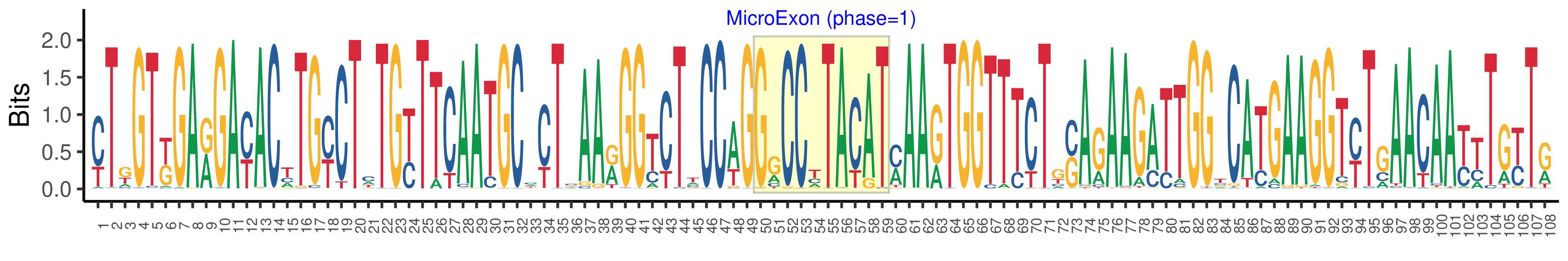 |
| Alignment of exons | 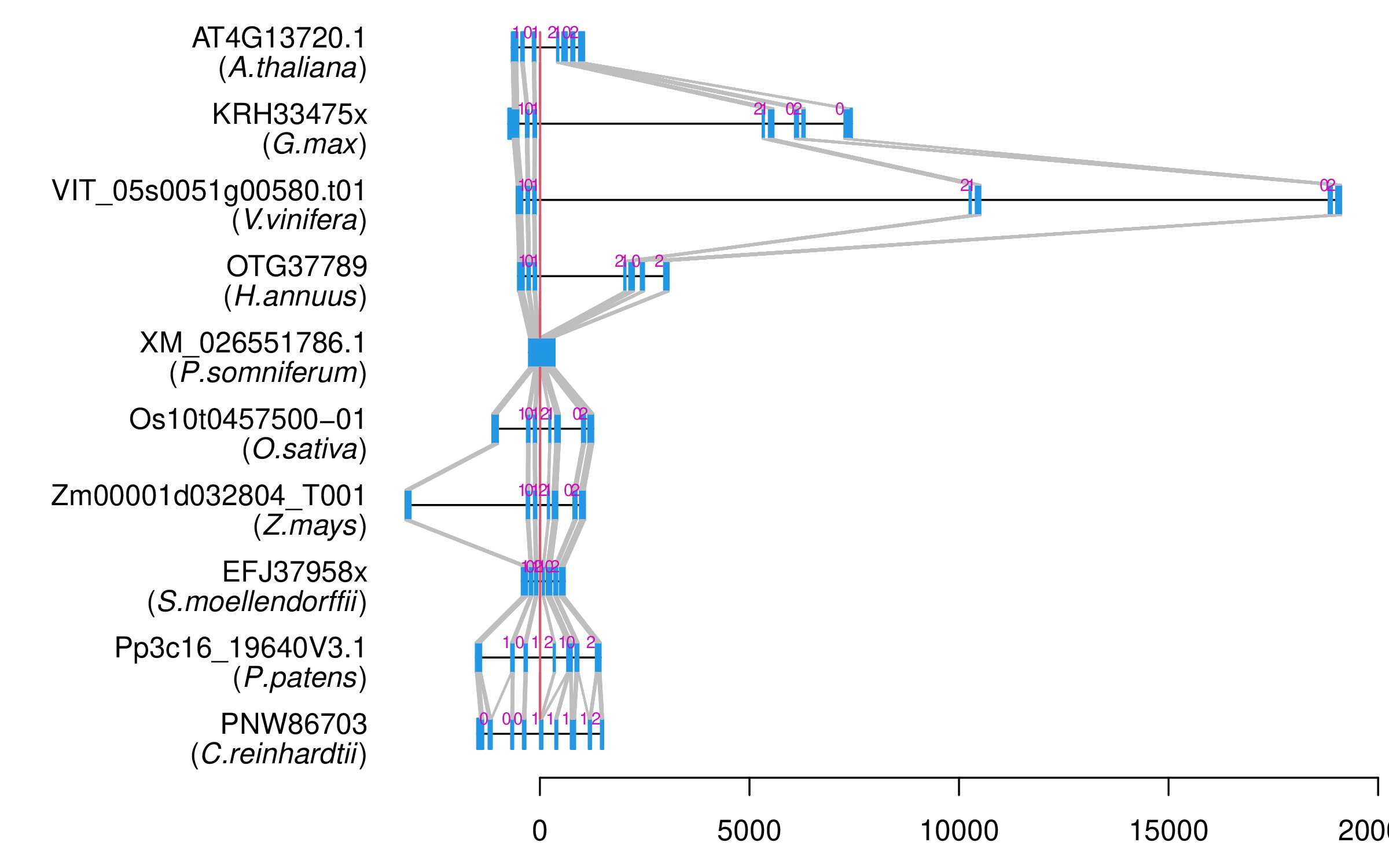 |
| Microexon DNA seq | GGCCTTATGT |
| Microexon Amino Acid seq | GPYV |
| Microexon-tag DNA Seq | CTCGTCGAGGATACATGCCTGTGCTTCAATGCTCTACATGGCCTTCCAGGGCCTTATGTGAAGTGGTTTCTTCAAAAGCTTGGCCACGAAGGCCTCAATAACATGCTG |
| Microexon-tag Amino Acid Seq | LVEDTCLCFNALHGLPGPYVKWFLQKLGHEGLNNML |
| Microexon-tag spanning region | 4057755-4058040 |
| Microexon-tag prediction score | 0.9408 |
| Overlapped with the annotated transcript (%) | 85.22 |
| New Transcript ID | EFJ37958x |
| Reference Transcript ID | EFJ37958 |
| Gene ID | SELMODRAFT_74723 |
| Gene Name | NA |
Sm_GL377565:4057862-4057871:+ does not have available information here.
| Microexon DNA seq | GGCCTTATGT |
| Microexon Amino Acid seq | GPYV |
| Microexon-tag DNA Seq | CTCGTCGAGGATACATGCCTGTGCTTCAATGCTCTACATGGCCTTCCAGGGCCTTATGTGAAGTGGTTTCTTCAAAAGCTTGGCCACGAAGGCCTCAATAACATGCTG |
| Microexon-tag Amino Acid seq | LVEDTCLCFNALHGLPGPYVKWFLQKLGHEGLNNML |
| Transcript ID | Sm.662.1 |
| Gene ID | Sm.662 |
| Gene Name | NA |
| Pfam domain motif | Ham1p_like |
| Motif E-value | 2.2e-54 |
| Motif start | 14 |
| Motif end | 188 |
| Protein seq | >Sm.662.1 MALVRAEVVLKKPVTFVTGNAKKLEEVKMILGNSIPFSTLRVDLPELQGEPEEISKEKARIAAKQIDGAVLVEDTCLCFN ALHGLPGPYVKWFLQKLGHEGLNNMLAAYKDKSAYALCVFSLALGPGFEPITFVGRTEGKIVPARGPADFGWDPVFQPDG SDFTYAEMPKDEKNKISHRRRALDKVRDHFREYDFVVRNDDS* |
| CDS seq | >Sm.662.1 ATGGCGCTAGTGAGGGCGGAGGTGGTGCTCAAGAAGCCCGTCACATTCGTCACAGGCAATGCCAAGAAGCTGGAGGAGGT AAAGATGATCCTCGGCAATTCCATCCCCTTCTCGACGCTGCGCGTTGATTTGCCGGAGCTTCAGGGAGAGCCCGAGGAGA TCTCGAAAGAAAAGGCGCGCATTGCTGCCAAACAGATTGATGGAGCTGTGCTCGTCGAGGATACATGCCTGTGCTTCAAT GCTCTACATGGCCTTCCAGGGCCTTATGTGAAGTGGTTTCTTCAAAAGCTTGGCCACGAAGGCCTCAATAACATGCTGGC AGCGTACAAAGATAAATCCGCTTATGCGTTGTGCGTCTTCTCACTAGCTCTCGGGCCTGGCTTTGAGCCTATAACATTTG TTGGACGCACTGAGGGGAAGATCGTTCCAGCGAGAGGGCCAGCAGATTTCGGCTGGGATCCGGTGTTCCAGCCAGATGGA AGCGATTTTACCTACGCAGAAATGCCCAAGGACGAGAAGAATAAGATCTCTCACCGGCGCCGCGCACTGGACAAAGTCCG AGATCATTTCCGGGAGTACGATTTCGTTGTGAGGAACGATGATTCATAA |