
| Microexon ID | Zm_2:58877082-58877090:+ |
| Species | Zea mays | Coordinates | 2:58877082..58877090 |
| Microexon Cluster ID | MEP22 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | Glyco_hydro_32N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | YSKYAMMGRACYGSYTWYCAYTTYCARCCYSMCAARAAYTGGATGAAYGATCCYAAYGGTCCAATGTWYTACAAGGGATKGTACCAYYTSTTCTAYCARTACAAYCCV |
| Logo of Microexon-tag DNA Seq | 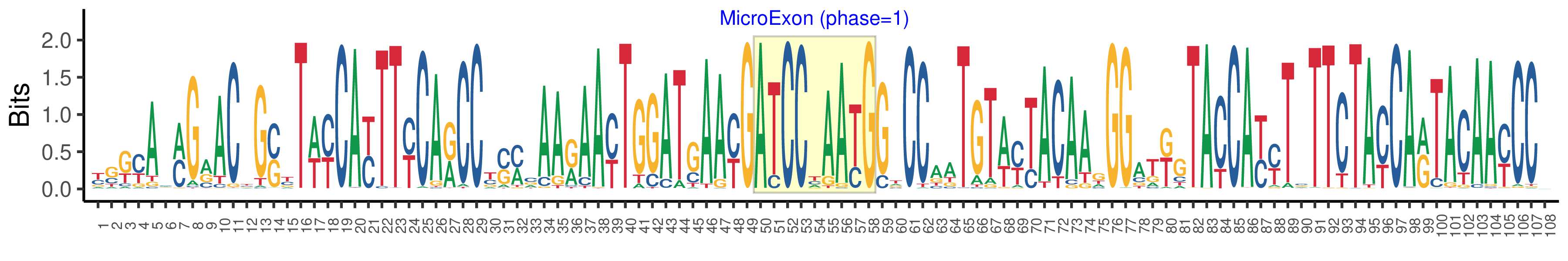 |
| Alignment of exons | 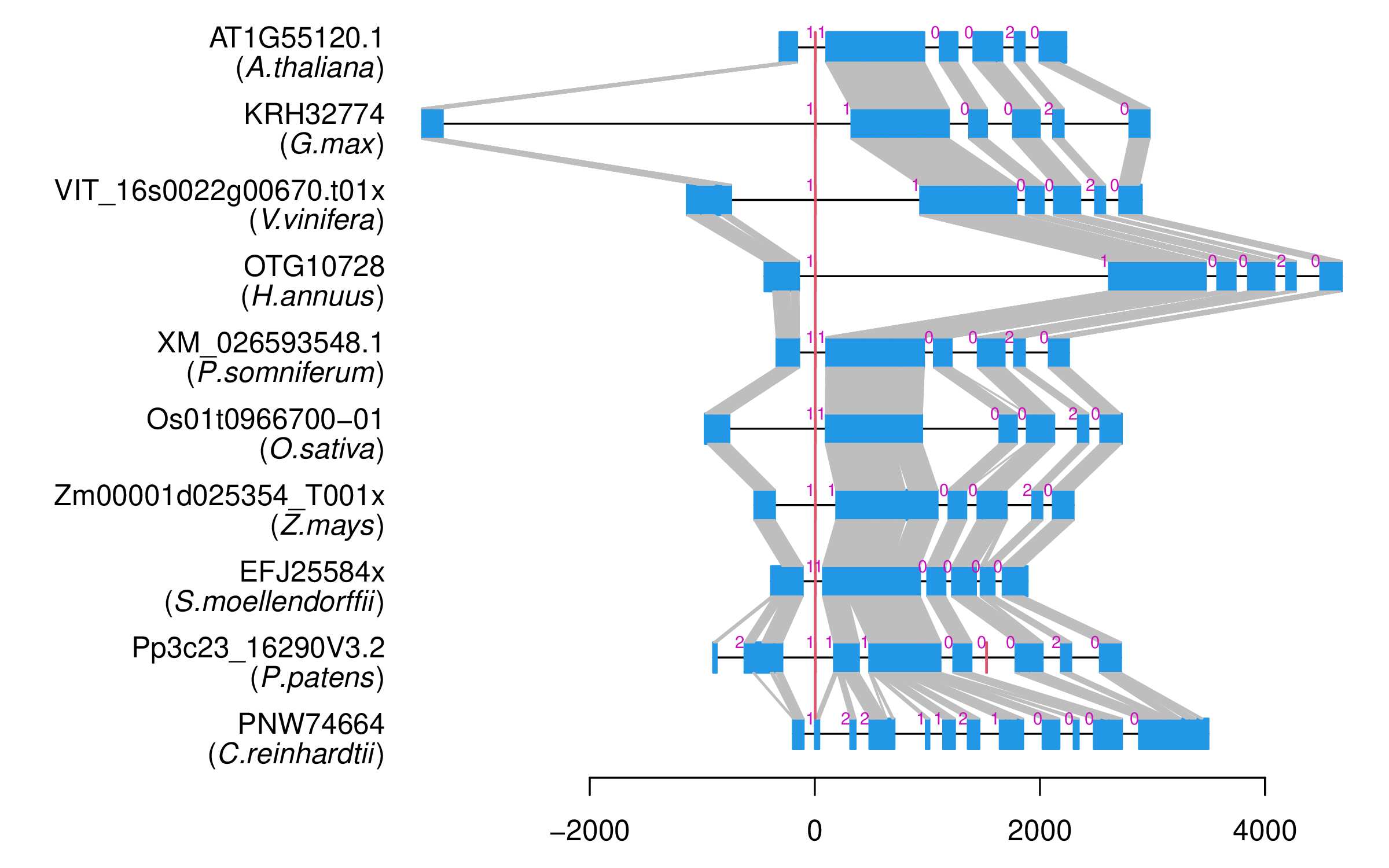 |
| Microexon DNA seq | ATCCCAACG |
| Microexon Amino Acid seq | DPNA |
| Microexon-tag DNA Seq | CTGCTCAGGACGGGGTACCACTTCCAGCCCCCCAAGAACTGGATCAATGATCCCAACGCGCCCATGTACTACAAGGGGTGGTACCATTTCTTCTACCAATACAATCCC |
| Microexon-tag Amino Acid Seq | MLRTGYHFQPPKNWINDPNAPMYYKGWYHFFYQYNP |
| Microexon-tag spanning region | 58876428-58877222 |
| Microexon-tag prediction score | 0.9732 |
| Overlapped with the annotated transcript (%) | 91.67 |
| New Transcript ID | Zm00001d003776_T001x |
| Reference Transcript ID | Zm00001d003776_T001 |
| Gene ID | Zm00001d003776 |
| Gene Name | miniature seed1 |
Zm_2:58877082-58877090:+ does not have available information here.
| Microexon DNA seq | ATCCCAACG |
| Microexon Amino Acid seq | DPNA |
| Microexon-tag DNA Seq | CTGCTCAGGACGGGGTACCACTTCCAGCCCCCCAAGAACTGGATCAATGATCCCAACGCGCCCATGTACTACAAGGGGTGGTACCATTTCTTCTACCAATACAATCCC |
| Microexon-tag Amino Acid seq | MLRTGYHFQPPKNWINDPNAPMYYKGWYHFFYQYNP |
| Transcript ID | Zm.10092.1 |
| Gene ID | Zm.10092 |
| Gene Name | miniature seed1 |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 5.6e-103 |
| Motif start | 58 |
| Motif end | 382 |
| Protein seq | >Zm.10092.1 MRALVVVSFASACLLLLLQLAGASHVVYNYKDLEAEAAAATDQVPPSIVNPLLRTGYHFQPPKNWINDPNAPMYYKGWYH FFYQYNPKGAVWGNIVWAHSVSRDLINWVALEPALRPSIPGDRYGCWSGSATVLPDGGGPVIMYTGVDHPDINYQVQNVA YPKNVSDPLLREWVKPSHNPVIVPEGGINATQFRDPTTAWRGPGPEQWRLLVGSAAGSSPRGVAYVYRSRDFRRWRRVRR PLHSAATGMWECPDFYPVSKGGAPRAGLETSVPPGPRVKHVLKNSLDLRRYDYYTVGTYHPRAERYVPDDPAGDEHRLRY DYGNFYASKTFYDPAKRRRILWGWANESDSAADDVAKGWAGIQAIPRTVWLDPSGKQLLQWPIEEVEALREKSVTLKNRL IKAGHHVEVTGIQTAQADVEVSFEVSPAALAGAETLDPALAYDAEKLCGVKRADVRGGVGPFGLWVLASANRKERTAVFF RVFKPAAGSDKPVVLMCTDPTKSSLNPNLYRPTFAGFVDTDISNGKISLRSLIDRSVVESFGAGGKTCILSRVYPSLAIG KDARLYVFNNGRAHVKVSRLTAWEMKKPVMNGA* |
| CDS seq | >Zm.10092.1 ATGAGAGCCCTCGTAGTCGTAAGTTTCGCTTCGGCGTGCTTGCTGCTGCTGTTGCAGCTCGCAGGAGCGTCGCATGTCGT CTACAACTACAAGGACCTCGAAGCCGAGGCTGCTGCGGCGACGGACCAGGTGCCGCCGTCCATCGTCAACCCCCTGCTCA GGACGGGGTACCACTTCCAGCCCCCCAAGAACTGGATCAATGATCCCAACGCGCCCATGTACTACAAGGGGTGGTACCAT TTCTTCTACCAATACAATCCCAAGGGCGCCGTATGGGGCAACATCGTGTGGGCGCACTCGGTGTCGCGAGACCTCATCAA CTGGGTGGCGCTAGAGCCGGCGCTCAGACCAAGCATCCCGGGCGACAGGTACGGCTGCTGGTCCGGGTCGGCGACGGTGC TGCCCGACGGCGGCGGCCCGGTGATCATGTACACGGGCGTGGACCACCCGGACATCAACTACCAGGTCCAGAACGTGGCG TACCCGAAGAACGTGTCGGACCCGCTGCTCCGGGAGTGGGTGAAGCCCTCGCACAACCCCGTCATCGTCCCCGAGGGCGG CATCAACGCGACGCAGTTCCGCGACCCGACCACGGCCTGGCGCGGCCCCGGCCCCGAGCAGTGGCGGCTGCTCGTGGGCA GCGCGGCGGGGTCGTCGCCCCGCGGCGTGGCGTACGTGTACCGCAGCCGCGACTTCCGGCGGTGGAGGCGGGTGCGGCGG CCGCTGCACTCGGCGGCCACGGGGATGTGGGAGTGCCCCGACTTCTACCCGGTGAGCAAGGGCGGCGCGCCGCGCGCGGG GCTCGAGACGTCCGTGCCGCCCGGCCCAAGGGTGAAGCACGTGCTCAAGAACAGCCTCGACCTCCGGCGGTACGACTACT ACACCGTGGGCACGTACCACCCGAGGGCCGAGCGGTACGTGCCGGACGACCCCGCCGGCGACGAGCACCGCCTGCGCTAC GACTACGGCAACTTCTACGCGTCCAAGACGTTCTACGACCCGGCCAAGCGGCGCCGCATCCTGTGGGGCTGGGCTAACGA GTCCGACTCCGCCGCCGACGATGTGGCCAAAGGCTGGGCTGGAATCCAGGCGATTCCAAGGACGGTGTGGCTGGACCCCA GCGGGAAGCAGCTGCTGCAGTGGCCTATCGAGGAGGTGGAGGCGCTGAGAGAAAAGTCGGTCACTCTCAAGAACAGGTTA ATCAAGGCAGGACATCACGTCGAGGTGACCGGGATACAAACGGCACAGGCTGACGTGGAGGTGAGCTTCGAGGTGTCGCC GGCGGCCCTGGCCGGTGCCGAGACGCTGGACCCGGCGCTGGCCTACGACGCGGAGAAGCTGTGCGGCGTGAAGCGCGCGG ACGTGAGGGGCGGCGTGGGGCCGTTCGGGCTGTGGGTGCTGGCCTCCGCCAACCGCAAGGAGAGAACCGCCGTGTTCTTC AGGGTGTTCAAGCCCGCCGCCGGCAGCGACAAGCCCGTGGTGCTCATGTGCACCGACCCTACCAAGTCGTCCCTGAACCC GAACCTGTACCGCCCGACATTTGCAGGATTTGTCGACACGGACATCTCGAACGGCAAGATATCCCTGAGAAGCCTGATCG ACCGGTCGGTCGTTGAGAGCTTCGGAGCCGGGGGCAAGACCTGCATCCTCTCCAGGGTCTACCCGTCGCTGGCCATCGGC AAGGACGCTCGCCTCTACGTGTTCAACAACGGGAGGGCGCACGTCAAGGTGTCCCGTCTCACCGCGTGGGAGATGAAGAA GCCGGTCATGAACGGGGCCTGA |