
| Microexon ID | Zm_10:115297369-115297377:+ |
| Species | Zea mays | Coordinates | 10:115297369..115297377 |
| Microexon Cluster ID | MEP22 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | Glyco_hydro_32N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | YSKYAMMGRACYGSYTWYCAYTTYCARCCYSMCAARAAYTGGATGAAYGATCCYAAYGGTCCAATGTWYTACAAGGGATKGTACCAYYTSTTCTAYCARTACAAYCCV |
| Logo of Microexon-tag DNA Seq | 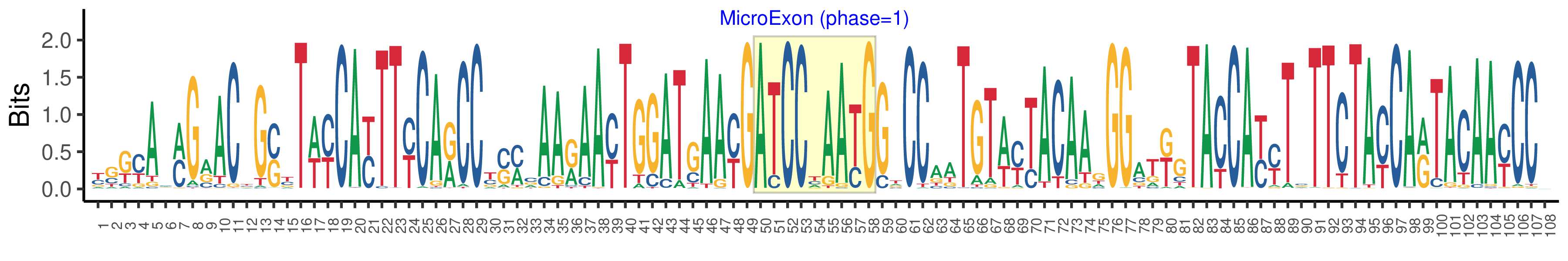 |
| Alignment of exons | 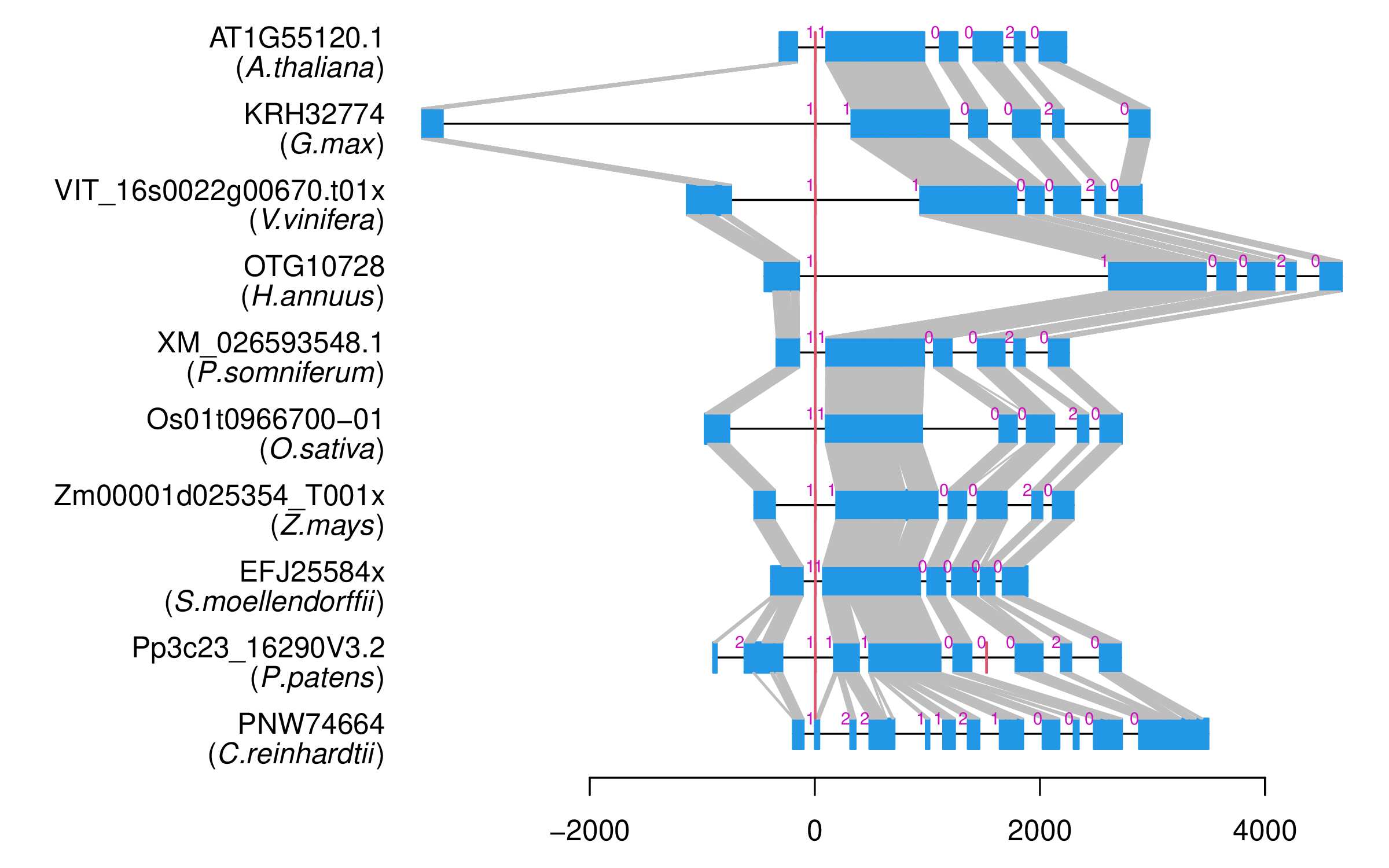 |
| Microexon DNA seq | ATCCAAATG |
| Microexon Amino Acid seq | DPNG |
| Microexon-tag DNA Seq | ACGTCGCGCACCGGGTACCATTTCCAGCCTCCTAAGCACTGGATCAACGATCCAAATGGACCAATGTACTACAAGGGGCTGTACCACCTGTTCTACCAGTACAACCCC |
| Microexon-tag Amino Acid Seq | TSRTGYHFQPPKHWINDPNGPMYYKGLYHLFYQYNP |
| Microexon-tag spanning region | 115297178-115297542 |
| Microexon-tag prediction score | 0.9676 |
| Overlapped with the annotated transcript (%) | 91.67 |
| New Transcript ID | Zm00001d025355_T001x |
| Reference Transcript ID | Zm00001d025355_T001 |
| Gene ID | Zm00001d025355 |
| Gene Name | invertase cell wall3 |
Zm_10:115297369-115297377:+ does not have available information here.
| Microexon DNA seq | ATCCAAATG |
| Microexon Amino Acid seq | DPNG |
| Microexon-tag DNA Seq | ACGTCGCGCACCGGGTACCATTTCCAGCCTCCTAAGCACTGGATCAACGATCCAAATGGACCAATGTACTACAAGGGGCTGTACCACCTGTTCTACCAGTACAACCCC |
| Microexon-tag Amino Acid seq | TSRTGYHFQPPKHWINDPNGPMYYKGLYHLFYQYNP |
| Transcript ID | Zm.7454.2 |
| Gene ID | Zm.7454 |
| Gene Name | invertase cell wall3 |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 3.6e-106 |
| Motif start | 48 |
| Motif end | 374 |
| Protein seq | >Zm.7454.2 MAALLLVWALLHCTGTAVTMVRASHAVYPELQSLEAEKVDETSRTGYHFQPPKHWINDPNGPMYYKGLYHLFYQYNPKGA VWGNIVWAHSVSTDLIDWTALAPGIYPSRQFDVKGCWSGSATVLPSGVPVVMYTGIDPDEHQVQNVAYPKNLSDPFLREW VKPDYNPIIAPDSGVNASAFRDPTTAWYGPDRHWRLLVGSKVGGKGLAVLYRSRDFRRWVKAHHPLHSGLTGMWECPDFF PVAVHGGGRHYRRGVDTAELHDRALAEEVKYVLKVSLDLTRYEYYTVGTYDHATDRYTPDAGFRDNDYGLRYDYGDFYAS KSFYDPAKRRRVLWGWANESDTVPDDRRKGWAGIQAIPRKLWLSPRGKQLIQWPVEEVKALRAKHVNVSDKVVKSGQYFE VTGFKSVQSDVEVEFAIGDLSKAERFNPRWLTDPQALCKKRGARETGEVGPFGLWVLAAGDLTERTAVFFRVFRANSNSS SSRLVVLMCNDPTNSTFEAQVYRPTFASFVNVDIARTKTIALRTLIDHSVVESFGAGGRTCILTRVYPKKALGDNAHLFV FNHGEVDVKVTRLDAWEMRTPKMNAPAQ* |
| CDS seq | >Zm.7454.2 ATGGCGGCGCTGTTGCTTGTGTGGGCTCTGCTGCACTGCACAGGCACGGCGGTGACGATGGTGCGGGCGTCCCACGCCGT GTATCCGGAGCTCCAGTCACTGGAGGCGGAGAAGGTTGATGAGACGTCGCGCACCGGGTACCATTTCCAGCCTCCTAAGC ACTGGATCAACGATCCAAATGGACCAATGTACTACAAGGGGCTGTACCACCTGTTCTACCAGTACAACCCCAAGGGCGCG GTGTGGGGCAACATCGTGTGGGCGCACTCGGTGTCGACCGACCTGATCGACTGGACGGCGCTGGCCCCGGGGATCTACCC GTCGAGGCAGTTCGACGTGAAGGGCTGCTGGTCGGGCTCCGCGACGGTGCTGCCGAGCGGCGTGCCGGTGGTGATGTACA CGGGCATCGACCCGGACGAGCACCAGGTGCAGAACGTGGCGTACCCCAAGAACCTGTCGGACCCGTTCCTCCGCGAGTGG GTGAAGCCCGACTACAACCCCATCATCGCCCCCGACAGCGGCGTGAACGCCAGCGCCTTCCGCGACCCGACCACCGCCTG GTACGGGCCCGACCGGCACTGGCGCCTGCTGGTGGGCAGCAAGGTGGGCGGCAAGGGCCTGGCCGTGCTGTACCGGAGCC GGGACTTCAGGCGCTGGGTCAAGGCGCACCACCCGCTCCACTCCGGCCTCACGGGGATGTGGGAGTGCCCGGACTTCTTC CCCGTCGCGGTGCACGGCGGCGGCCGCCACTACCGCCGCGGCGTCGACACCGCCGAGCTGCACGACAGGGCCCTGGCCGA GGAGGTCAAGTACGTGCTCAAGGTCAGCCTGGACCTCACGCGCTACGAGTACTACACGGTGGGCACCTACGACCACGCCA CGGACCGCTACACCCCCGACGCCGGCTTCCGCGACAACGACTACGGCCTGCGCTACGACTACGGAGACTTCTACGCGTCC AAGTCCTTCTACGATCCGGCCAAGCGCCGCCGCGTGCTGTGGGGCTGGGCCAACGAGTCGGACACCGTTCCTGATGACCG CAGGAAGGGCTGGGCCGGGATTCAGGCGATACCGAGGAAGCTGTGGCTGTCGCCGCGCGGGAAGCAGCTGATCCAGTGGC CGGTGGAGGAGGTCAAGGCTCTGCGCGCCAAGCACGTGAACGTCAGTGACAAGGTCGTCAAGAGCGGGCAGTACTTCGAG GTCACCGGCTTCAAGTCCGTGCAGTCCGACGTGGAGGTGGAGTTCGCGATCGGCGACCTAAGCAAGGCGGAGCGGTTCAA CCCCAGGTGGCTGACCGACCCGCAGGCGCTGTGCAAGAAGCGGGGCGCCCGGGAGACGGGCGAGGTGGGGCCGTTCGGGC TGTGGGTGCTGGCCGCTGGCGACCTCACGGAGAGGACGGCCGTCTTCTTCAGGGTGTTCCGGGCCAACAGCAACAGCAGC AGCAGCAGGCTCGTCGTCCTCATGTGCAACGACCCTACTAACTCCACCTTCGAGGCGCAGGTCTACAGGCCCACCTTCGC CAGCTTCGTCAACGTCGACATCGCCAGAACCAAGACGATCGCGCTGAGGACACTGATCGACCACTCCGTGGTGGAGAGCT TCGGCGCGGGCGGCAGGACGTGCATCCTGACGAGGGTGTACCCCAAGAAGGCCCTGGGCGACAACGCGCACCTCTTCGTC TTCAACCACGGCGAGGTGGACGTCAAGGTCACCAGGCTGGACGCCTGGGAGATGAGGACCCCGAAGATGAACGCGCCGGC GCAGTAG |