
| Microexon ID | Vv_16:12050861-12050869:+ |
| Species | Vistis vinifera | Coordinates | 16:12050861..12050869 |
| Microexon Cluster ID | MEP22 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | Glyco_hydro_32N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | YSKYAMMGRACYGSYTWYCAYTTYCARCCYSMCAARAAYTGGATGAAYGATCCYAAYGGTCCAATGTWYTACAAGGGATKGTACCAYYTSTTCTAYCARTACAAYCCV |
| Logo of Microexon-tag DNA Seq | 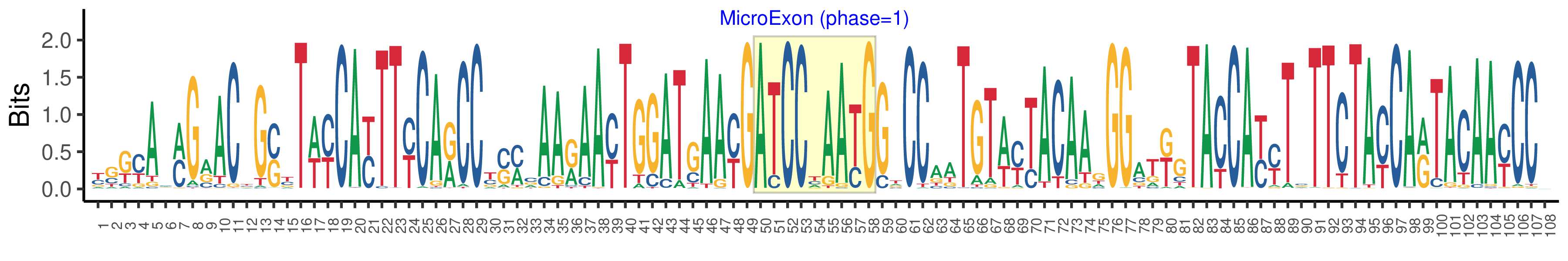 |
| Alignment of exons | 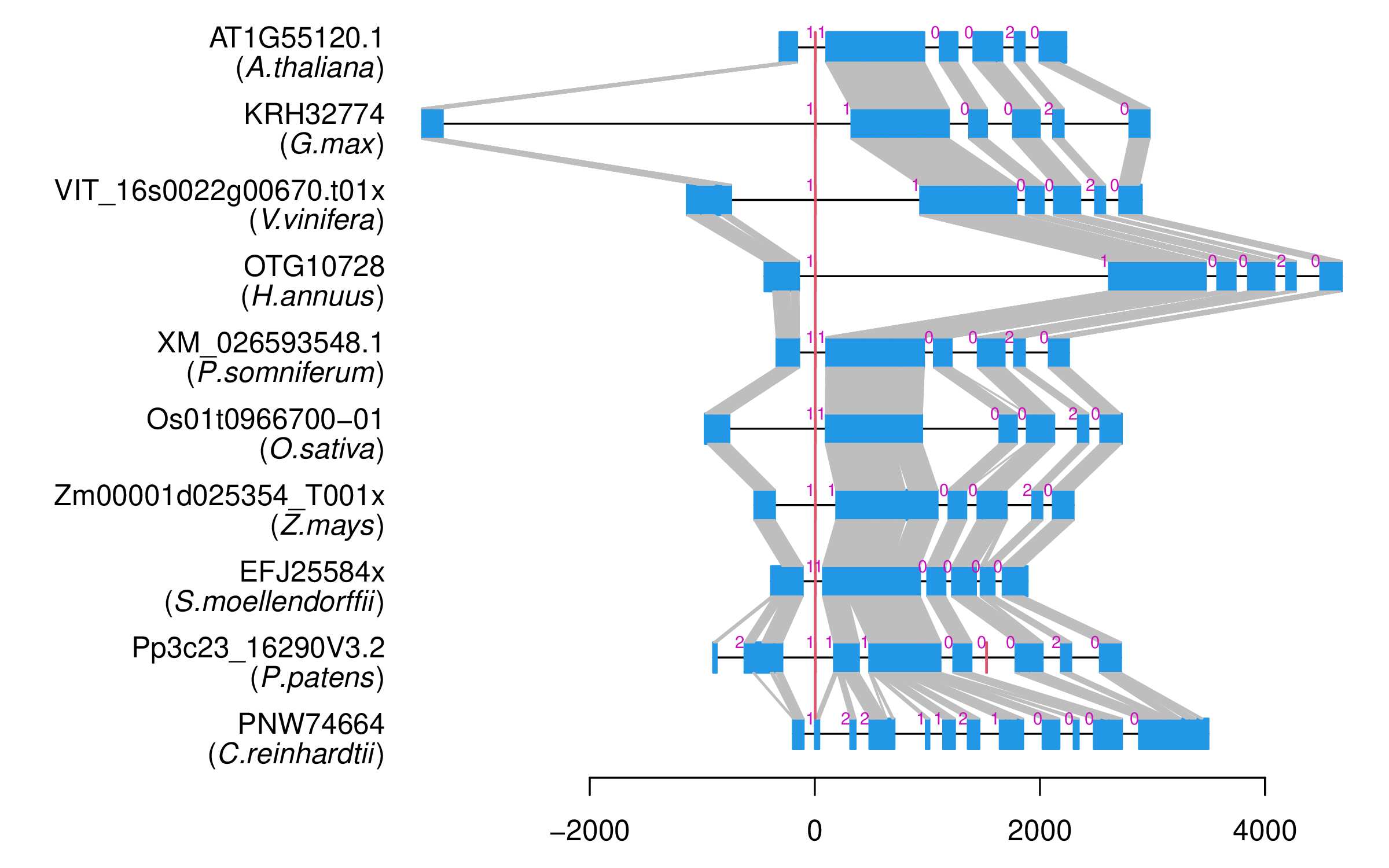 |
| Microexon DNA seq | ATCCTGACG |
| Microexon Amino Acid seq | DPDG |
| Microexon-tag DNA Seq | TGGCAGAGAACTGCCTTCCATTTCCAGCCTGAGAAGAATTGGATGAATGATCCTGACGGACCACTGTTCCACATGGGATGGTACCACCTCTTCTACCAGTACAATCCT |
| Microexon-tag Amino Acid Seq | WQRTAFHFQPEKNWMNDPDGPLFHMGWYHLFYQYNP |
| Microexon-tag spanning region | 12050069-12051846 |
| Microexon-tag prediction score | 0.9732 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | VIT_16s0022g00670.t01x |
| Reference Transcript ID | VIT_16s0022g00670.t01 |
| Gene ID | VIT_16s0022g00670 |
| Gene Name | NA |
| Transcript ID | VIT_16s0022g00670.t01 |
| Protein ID | VIT_16s0022g00670.t01 |
| Gene ID | VIT_16s0022g00670 |
| Gene Name | NA |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 1.3e-100 |
| Motif start | 10 |
| Motif end | 328 |
| Protein seq | >VIT_16s0022g00670.t01 MFAWQRTAFHFQPEKNWMNDPDGPLFHMGWYHLFYQYNPDSAVWGNITWGHAVSRDMIHWLYLPLAMVPDRWFDLNGVWT GSATILPNGQIIMLYTGDTNDSVQVQNLAYPANLSDPLLLHWIKYENNPVMVPPAGIGSDDFRDPTTMWVGADGNWRVAV GSLVNTTGIVLVFQTTNFTDFELLDGELHGVPGTGMWECVDFYPVSINGVYGLDTSAHGPGIKHVLKASMDDNRHDYYAL GEYDPMTDTWTPDDPELDVGIGLRLDYGKYYASKTFYDQVKKRRILYGWISEGDIESDDLKKGWASLQSIPRTVLHDNKT GTYLLLWPIEEVESLRTNSTEFEDVLLEPGSVVPLDIGSASQLDIVAEFEVDNETLEAMVEADVIYNCSTSAGAAGRGAL GPFGILVLADDTLSELTPIYFYIAKDTDGSYKTFFCTDLSRSSLAVDDVDQRIYGSIVPVLDDEKPTMRVLVDHSIVEGF SQGGRSCITTRVYPTEAIYGAARLFLFNNATGVNVTASIKIWEMASADIHPYPLDQP* |
| CDS seq | >VIT_16s0022g00670.t01 ATGTTTGCCTGGCAGAGAACTGCCTTCCATTTCCAGCCTGAGAAGAATTGGATGAATGATCCTGACGGACCACTGTTCCA CATGGGATGGTACCACCTCTTCTACCAGTACAATCCTGATTCAGCAGTGTGGGGGAACATCACGTGGGGCCATGCAGTGT CAAGAGACATGATCCACTGGCTCTACCTCCCCTTAGCCATGGTCCCCGATCGATGGTTTGATTTGAACGGCGTCTGGACC GGCTCTGCTACGATTCTGCCAAATGGCCAAATCATAATGCTCTACACCGGTGATACCAATGATTCTGTGCAGGTCCAAAA TCTAGCATACCCTGCCAACCTGTCTGATCCCCTCCTCCTCCATTGGATCAAATATGAAAACAATCCGGTTATGGTCCCCC CAGCCGGAATCGGTTCCGACGATTTCCGGGACCCGACCACGATGTGGGTCGGAGCCGATGGAAATTGGCGGGTTGCAGTT GGGTCACTGGTCAATACAACAGGCATCGTGCTTGTATTTCAGACCACCAACTTTACCGATTTTGAGCTGTTGGACGGAGA ATTGCATGGGGTTCCGGGTACGGGTATGTGGGAGTGCGTGGACTTTTACCCGGTTTCCATCAATGGCGTATACGGGTTGG ATACGTCGGCTCATGGGCCGGGCATAAAGCATGTGTTGAAGGCCAGCATGGATGATAACAGGCATGATTATTACGCATTA GGGGAGTATGACCCGATGACCGATACATGGACACCCGATGACCCGGAATTGGATGTGGGTATCGGGTTACGGCTTGATTA TGGAAAGTATTATGCTTCAAAGACATTTTATGATCAGGTTAAGAAAAGGAGGATCCTATATGGTTGGATTTCGGAAGGTG ATATTGAATCTGATGACTTGAAGAAAGGATGGGCTTCCCTTCAGAGTATACCAAGGACTGTTCTACATGACAATAAGACA GGAACCTATCTCCTTCTATGGCCAATTGAGGAGGTAGAGAGCTTGAGAACAAACAGTACAGAGTTCGAAGATGTTTTACT CGAACCCGGATCTGTTGTGCCACTTGACATAGGCTCAGCTTCACAGTTGGACATAGTGGCGGAGTTTGAGGTAGATAACG AGACCTTAGAAGCGATGGTGGAAGCTGATGTGATCTATAATTGTAGTACTAGCGCTGGTGCTGCTGGGAGGGGAGCCTTG GGGCCATTCGGCATTCTGGTTTTAGCAGATGATACACTTTCCGAGCTGACCCCAATTTATTTTTACATTGCCAAAGACAC TGATGGCAGTTACAAAACCTTCTTCTGCACTGATCTATCCAGGTCGTCTTTGGCCGTGGATGATGTTGATCAAAGGATTT ATGGGAGCATTGTTCCAGTGCTTGATGATGAAAAACCAACCATGAGGGTTCTGGTTGATCATTCGATTGTGGAAGGCTTT AGCCAAGGGGGGAGGTCATGTATCACGACCCGAGTTTATCCAACAGAGGCTATCTATGGAGCAGCTAGGTTATTCCTCTT CAACAATGCCACTGGAGTGAATGTCACAGCCAGTATCAAGATATGGGAAATGGCTTCTGCTGATATTCACCCTTACCCAT TAGACCAACCATAA |
| Microexon DNA seq | ATCCTGACG |
| Microexon Amino Acid seq | DPDG |
| Microexon-tag DNA Seq | TGGCAGAGAACTGCCTTCCATTTCCAGCCTGAGAAGAATTGGATGAATGATCCTGACGGACCACTGTTCCACATGGGATGGTACCACCTCTTCTACCAGTACAATCCT |
| Microexon-tag Amino Acid seq | WQRTAFHFQPEKNWMNDPDGPLFHMGWYHLFYQYNP |
| Transcript ID | VIT_16s0022g00670.t01 |
| Gene ID | Vv.12737 |
| Gene Name | NA |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 2.2e-100 |
| Motif start | 122 |
| Motif end | 440 |
| Protein seq | >VIT_16s0022g00670.t01 MFAWQRTAFHFQPEKNWMNDPDGPLFHMGWYHLFYQYNPDSAVWGNITWGHAVSRDMIHWLYLPLAMVPDRWFDLNGVWT GSATILPNGQIIMLYTGDTNDSVQVQNLAYPANLSDPLLLHWIKYENNPVMVPPAGIGSDDFRDPTTMWVGADGNWRVAV GSLVNTTGIVLVFQTTNFTDFELLDGELHGVPGTGMWECVDFYPVSINGVYGLDTSAHGPGIKHVLKASMDDNRHDYYAL GEYDPMTDTWTPDDPELDVGIGLRLDYGKYYASKTFYDQVKKRRILYGWISEGDIESDDLKKGWASLQSIPRTVLHDNKT GTYLLLWPIEEVESLRTNSTEFEDVLLEPGSVVPLDIGSASQLDIVAEFEVDNETLEAMVEADVIYNCSTSAGAAGRGAL GPFGILVLADDTLSELTPIYFYIAKDTDGSYKTFFCTDLSRSSLAVDDVDQRIYGSIVPVLDDEKPTMRVLVDHSIVEGF SQGGRSCITTRVYPTEAIYGAARLFLFNNATGVNVTASIKIWEMASADIHPYPLDQP* |
| CDS seq | >VIT_16s0022g00670.t01 ATGTTTGCCTGGCAGAGAACTGCCTTCCATTTCCAGCCTGAGAAGAATTGGATGAATGATCCTGACGGACCACTGTTCCA CATGGGATGGTACCACCTCTTCTACCAGTACAATCCTGATTCAGCAGTGTGGGGGAACATCACGTGGGGCCATGCAGTGT CAAGAGACATGATCCACTGGCTCTACCTCCCCTTAGCCATGGTCCCCGATCGATGGTTTGATTTGAACGGCGTCTGGACC GGCTCTGCTACGATTCTGCCAAATGGCCAAATCATAATGCTCTACACCGGTGATACCAATGATTCTGTGCAGGTCCAAAA TCTAGCATACCCTGCCAACCTGTCTGATCCCCTCCTCCTCCATTGGATCAAATATGAAAACAATCCGGTTATGGTCCCCC CAGCCGGAATCGGTTCCGACGATTTCCGGGACCCGACCACGATGTGGGTCGGAGCCGATGGAAATTGGCGGGTTGCAGTT GGGTCACTGGTCAATACAACAGGCATCGTGCTTGTATTTCAGACCACCAACTTTACCGATTTTGAGCTGTTGGACGGAGA ATTGCATGGGGTTCCGGGTACGGGTATGTGGGAGTGCGTGGACTTTTACCCGGTTTCCATCAATGGCGTATACGGGTTGG ATACGTCGGCTCATGGGCCGGGCATAAAGCATGTGTTGAAGGCCAGCATGGATGATAACAGGCATGATTATTACGCATTA GGGGAGTATGACCCGATGACCGATACATGGACACCCGATGACCCGGAATTGGATGTGGGTATCGGGTTACGGCTTGATTA TGGAAAGTATTATGCTTCAAAGACATTTTATGATCAGGTTAAGAAAAGGAGGATCCTATATGGTTGGATTTCGGAAGGTG ATATTGAATCTGATGACTTGAAGAAAGGATGGGCTTCCCTTCAGAGTATACCAAGGACTGTTCTACATGACAATAAGACA GGAACCTATCTCCTTCTATGGCCAATTGAGGAGGTAGAGAGCTTGAGAACAAACAGTACAGAGTTCGAAGATGTTTTACT CGAACCCGGATCTGTTGTGCCACTTGACATAGGCTCAGCTTCACAGTTGGACATAGTGGCGGAGTTTGAGGTAGATAACG AGACCTTAGAAGCGATGGTGGAAGCTGATGTGATCTATAATTGTAGTACTAGCGCTGGTGCTGCTGGGAGGGGAGCCTTG GGGCCATTCGGCATTCTGGTTTTAGCAGATGATACACTTTCCGAGCTGACCCCAATTTATTTTTACATTGCCAAAGACAC TGATGGCAGTTACAAAACCTTCTTCTGCACTGATCTATCCAGGTCGTCTTTGGCCGTGGATGATGTTGATCAAAGGATTT ATGGGAGCATTGTTCCAGTGCTTGATGATGAAAAACCAACCATGAGGGTTCTGGTTGATCATTCGATTGTGGAAGGCTTT AGCCAAGGGGGGAGGTCATGTATCACGACCCGAGTTTATCCAACAGAGGCTATCTATGGAGCAGCTAGGTTATTCCTCTT CAACAATGCCACTGGAGTGAATGTCACAGCCAGTATCAAGATATGGGAAATGGCTTCTGCTGATATTCACCCTTACCCAT TAGACCAACCATAA |