
| Microexon ID | Ta_7A:4016629-4016637:- |
| Species | Triticum Aestivum | Coordinates | 7A:4016629..4016637 |
| Microexon Cluster ID | MEP22 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | Glyco_hydro_32N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | YSKYAMMGRACYGSYTWYCAYTTYCARCCYSMCAARAAYTGGATGAAYGATCCYAAYGGTCCAATGTWYTACAAGGGATKGTACCAYYTSTTCTAYCARTACAAYCCV |
| Logo of Microexon-tag DNA Seq | 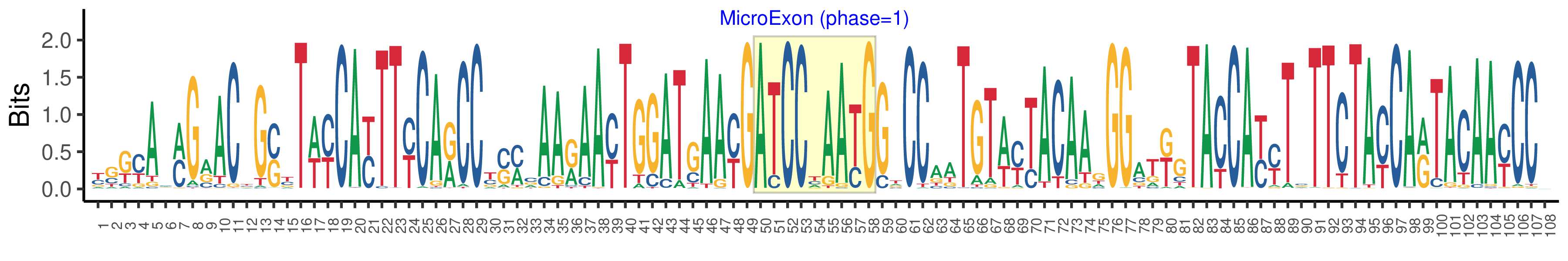 |
| Alignment of exons |  |
| Microexon DNA seq | ATCCCAACG |
| Microexon Amino Acid seq | DPNG |
| Microexon-tag DNA Seq | TGGCAGCGCACCGGCTACCATTTCCAGCCGGAGAAGAACTACCAGAACGATCCCAACGGTCCGGTGTACTACAAAGGATGGTACCACTTCTTCTACCAGCACAACCCG |
| Microexon-tag Amino Acid Seq | WQRTGYHFQPEKNYQNDPNGPVYYKGWYHFFYQHNP |
| Microexon-tag spanning region | 4016200-4016842 |
| Microexon-tag prediction score | 0.9642 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | TraesCS7A02G009200.2x |
| Reference Transcript ID | TraesCS7A02G009200.2 |
| Gene ID | TraesCS7A02G009200 |
| Gene Name | NA |
| Transcript ID | TraesCS7A02G009200.2 |
| Protein ID | |
| Gene ID | TraesCS7A02G009200 |
| Gene Name | |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 3.00E-88 |
| Motif start | 113 |
| Motif end | 435 |
| Protein seq | >TraesCS7A02G009200.2 MESSRGILIPGTPPLPYAYEPLPSSSADANGQEDRRITGGVRWRAWAAVLAVGALVVAAAVFGASRVDRDAVASSVPATA EHGVLEKASGPYSASGGFPWSNAMLQWQRTGYHFQPEKNYQNDPNGPVYYKGWYHFFYQHNPGGTGWGNISWGHAVSRDM VHWRHLPLAMVPEHWYDIEGVLTGSITVLPDGRVILLYTGNTETFAQVTCLAEAAEPSDPLLREWVKHPANPVVYPPPGI GMKDYRDPTTAWFDNSDNTWRIIIGSKNDTDHSGIVFTYKTKDFVSYELIPGYLYRGPAGTGMYECIDLFAVGGGRAASD MYNSTAEDVLYVLKESSDDDRRDYYALGRFDAAANTWTPIDTEQELGVALRYDYGRYDASKSFYDPVKQRRIVWGYVVET DSWSADAAKGWANLQSIPRTVALDEKTRTNLIQWPVEELDTLRINTTDLSGISVGAGSVVSLPLHQTSQLDIEASFRINA STIEALNEVDVGYNCTMTSGAATRGALGPFGILVLANVALTEQTAVYFYVSKGLDGGLRTHFCHDELRSTHATDVAKEVV GSTVPVLDGEDFSVRVLVDHSIVQSFVMGGRMTATSRAYPTEAIYAAAGVYLFNNATGASITAEKLVVHDMDSSYNRIFT DEDLLVLD* |
| CDS seq | >TraesCS7A02G009200.2 ATGGAGTCGTCACGCGGCATCCTCATCCCCGGCACGCCGCCGCTGCCCTACGCGTACGAGCCGCTGCCCTCCTCCTCCGC CGACGCCAACGGCCAGGAGGACCGGCGGATCACCGGCGGCGTGAGGTGGCGCGCGTGGGCCGCCGTGCTGGCCGTGGGGG CGCTCGTCGTCGCCGCCGCGGTCTTTGGGGCCAGCAGGGTCGACCGGGACGCGGTGGCCTCCTCCGTACCGGCTACAGCG GAACATGGCGTGCTGGAGAAGGCGTCCGGGCCATACTCCGCCAGCGGCGGCTTCCCGTGGAGCAACGCCATGCTGCAGTG GCAGCGCACCGGCTACCATTTCCAGCCGGAGAAGAACTACCAGAACGATCCCAACGGTCCGGTGTACTACAAAGGATGGT ACCACTTCTTCTACCAGCACAACCCGGGGGGCACCGGGTGGGGCAACATCTCATGGGGCCATGCCGTGTCGCGGGACATG GTCCACTGGCGTCACCTACCGCTCGCCATGGTGCCCGAGCACTGGTACGACATCGAGGGTGTCCTCACCGGATCCATCAC CGTGCTCCCCGATGGCAGGGTCATCCTGCTCTACACAGGGAACACCGAGACGTTCGCGCAGGTGACCTGCCTCGCGGAGG CCGCTGAACCAAGCGACCCCCTCCTCCGCGAGTGGGTCAAGCACCCCGCCAACCCCGTCGTGTACCCGCCCCCTGGCATC GGCATGAAGGACTACCGTGACCCAACCACGGCATGGTTCGACAACTCTGACAACACGTGGCGCATCATCATTGGCTCCAA GAACGACACCGATCACTCCGGCATCGTCTTCACGTACAAGACCAAAGACTTCGTCAGCTACGAGCTGATACCGGGATACT TGTACCGTGGCCCCGCCGGCACCGGCATGTACGAGTGCATCGATCTATTTGCCGTGGGCGGCGGTCGCGCCGCCAGTGAC ATGTACAATTCGACGGCCGAGGACGTGCTGTACGTGCTCAAGGAGAGCAGCGACGACGACCGGCGCGACTATTACGCGCT CGGGAGGTTCGACGCAGCGGCCAACACATGGACGCCGATCGACACCGAGCAGGAACTCGGCGTCGCGCTGCGGTACGACT ATGGCAGGTACGACGCGTCCAAGTCCTTCTACGACCCCGTGAAGCAGCGGCGGATCGTCTGGGGGTACGTCGTCGAGACC GACTCCTGGAGCGCCGACGCCGCCAAGGGGTGGGCCAACCTCCAGTCGATTCCGAGGACGGTGGCGCTGGACGAGAAGAC CCGGACGAACCTCATCCAATGGCCGGTGGAGGAGCTCGATACCCTCCGCATCAACACCACCGATCTTAGCGGCATCAGTG TCGGTGCTGGATCCGTCGTCTCCCTCCCTCTCCACCAGACCTCCCAACTCGACATCGAGGCATCCTTCCGCATCAACGCC TCCACCATTGAGGCACTCAACGAGGTCGACGTCGGCTACAACTGCACCATGACCAGCGGCGCCGCCACCCGTGGCGCGCT CGGCCCCTTTGGCATTCTCGTTCTTGCCAACGTCGCCCTGACAGAACAGACGGCGGTGTACTTTTATGTGTCCAAGGGCC TCGACGGTGGTCTTCGGACGCACTTCTGCCACGATGAGTTGCGGTCGACACATGCTACCGATGTGGCGAAGGAGGTAGTT GGTAGCACGGTACCGGTGCTCGACGGCGAGGATTTTTCCGTTAGGGTGCTCGTGGACCACTCCATCGTGCAGAGCTTCGT GATGGGCGGGAGGATGACGGCGACGTCAAGGGCCTACCCGACGGAGGCCATTTACGCGGCGGCGGGGGTCTACCTGTTCA ACAATGCCACCGGCGCCAGCATCACCGCTGAGAAGCTCGTCGTGCACGACATGGATTCGTCGTACAACCGGATATTCACA GACGAAGACTTGTTAGTCCTCGATTAG |
Ta_7A:4016629-4016637:- does not have available information here.