
| Microexon ID | Ta_4A:739415986-739415994:+ |
| Species | Triticum Aestivum | Coordinates | 4A:739415986..739415994 |
| Microexon Cluster ID | MEP22 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | Glyco_hydro_32N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | YSKYAMMGRACYGSYTWYCAYTTYCARCCYSMCAARAAYTGGATGAAYGATCCYAAYGGTCCAATGTWYTACAAGGGATKGTACCAYYTSTTCTAYCARTACAAYCCV |
| Logo of Microexon-tag DNA Seq | 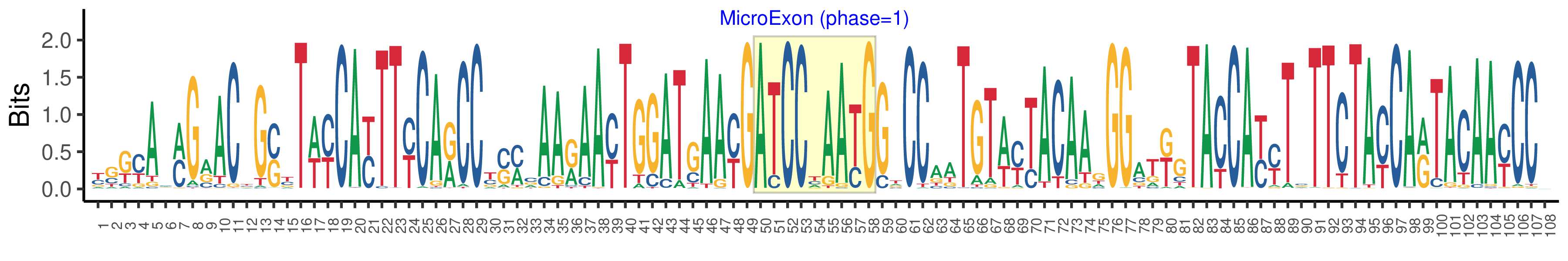 |
| Alignment of exons |  |
| Microexon DNA seq | ATCCCAACG |
| Microexon Amino Acid seq | DPNG |
| Microexon-tag DNA Seq | TGGCAGCGCACCGGCTACCATTTCCAGCCGGACAAGTACTACCAGAACGATCCCAACGGTCCGGTGTACTACGGCGGATGGTACCACTTCTTCTACCAATACAACCCG |
| Microexon-tag Amino Acid Seq | WQRTGYHFQPDKYYQNDPNGPVYYGGWYHFFYQYNP |
| Microexon-tag spanning region | 739415773-739416158 |
| Microexon-tag prediction score | 0.9527 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | TraesCS4A02G486000.1x |
| Reference Transcript ID | TraesCS4A02G486000.1 |
| Gene ID | TraesCS4A02G486000 |
| Gene Name | NA |
| Transcript ID | TraesCS4A02G486000.1 |
| Protein ID | |
| Gene ID | TraesCS4A02G486000 |
| Gene Name | |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 6.20E-89 |
| Motif start | 128 |
| Motif end | 451 |
| Protein seq | >TraesCS4A02G486000.1 MDSSRVILIPGTPPLPYAAYEQLPSSSADAKQGQEEQRAGGGLRWRACAAVLAASAVVALVVAAAVFGAGGAGRDAVAAS VPATSATGFPRSRGKEHGVSEKTSGAYSANGGFPWSNAMLQWQRTGYHFQPDKYYQNDPNGPVYYGGWYHFFYQYNPSGS VWEPQIVWGHAVSKDLIHWRHLPPALVPDQWYDIKGVLTGSITVLPDGKVVLLYTGNTETFAQVTCLAEPADPSDPLLRE WVKHPANPVVFPPPGIGMKDFRDPTTAWFDESDGTWRTIIGSKNDSDHSGIVFSYKTKDFVSYELMPGYMYRGPKGTGEY ECIDLYAVGGGRKASDMYNSTAEDVLYVLKESSDDDRHDWYSLGRFDAAANKWTPIDTELELGVGLRYDWGKYYASKSFY DPVKQRRVVWAYVGETDSERADITKGWANLQSIPRTVELDEKTRTNLIQWPVEELDTLRINTTDLSGFTVGAGSVVPLHL HQTAQLDIEATFRIDASAIEALNEADVSYNCTTSSGAATRGALGPFGLLVLANRALTEQTGVYFYVSKGLDGGLRTHFCH DELRSSHASDVVKRVVGSTVPVLDGEDFSVRVLVDHSIVQSFAMGGRLTATSRAYPTEAIYAAAGVYMFNNATGTSVTAE KLVVHDMDSSYNHIYTDDDLVVVN* |
| CDS seq | >TraesCS4A02G486000.1 ATGGACTCGTCTCGCGTCATACTCATCCCCGGCACGCCGCCGCTGCCGTACGCCGCCTACGAGCAGCTGCCGTCCTCCTC CGCCGACGCCAAGCAGGGCCAGGAGGAGCAGCGGGCCGGCGGTGGCCTGAGGTGGCGCGCGTGCGCCGCCGTCCTGGCCG CCTCGGCCGTGGTGGCGCTCGTCGTGGCCGCCGCGGTCTTCGGCGCCGGCGGGGCGGGCCGTGACGCGGTGGCCGCCTCC GTGCCGGCGACCTCGGCGACGGGGTTCCCGAGGAGCAGGGGCAAGGAGCACGGCGTGTCGGAGAAGACGTCGGGGGCCTA CTCCGCCAACGGCGGGTTCCCGTGGAGCAACGCCATGCTGCAGTGGCAGCGCACCGGCTACCATTTCCAGCCGGACAAGT ACTACCAGAACGATCCCAACGGTCCGGTGTACTACGGCGGATGGTACCACTTCTTCTACCAATACAACCCGTCGGGCTCC GTGTGGGAGCCCCAGATCGTGTGGGGCCACGCCGTGTCCAAGGACCTCATCCACTGGCGCCACCTCCCGCCGGCCCTGGT GCCCGACCAGTGGTACGACATCAAGGGCGTCCTCACCGGCTCCATCACCGTGCTCCCCGACGGCAAGGTCGTCCTCCTCT ACACGGGCAACACCGAGACGTTCGCGCAGGTGACCTGCCTCGCGGAGCCCGCCGACCCGAGCGACCCGCTCCTCCGTGAG TGGGTGAAGCACCCCGCCAACCCCGTCGTGTTCCCGCCCCCCGGCATCGGCATGAAGGACTTCCGCGACCCCACCACGGC CTGGTTCGACGAGTCGGACGGCACGTGGCGCACCATCATCGGCTCCAAGAACGACTCGGACCACTCCGGCATCGTCTTCT CCTACAAGACCAAGGACTTCGTCAGCTACGAGCTGATGCCCGGGTACATGTACCGCGGCCCCAAGGGCACCGGCGAGTAC GAGTGCATCGACCTCTACGCCGTCGGCGGGGGCCGCAAGGCCAGCGACATGTACAACTCGACGGCCGAGGACGTGCTGTA CGTGCTCAAGGAGAGCAGCGACGACGACCGGCACGACTGGTACTCGCTGGGCCGGTTCGACGCCGCCGCCAACAAGTGGA CGCCGATCGACACGGAGCTGGAGCTCGGCGTCGGGCTCCGGTACGACTGGGGCAAGTACTACGCGTCCAAGTCCTTCTAC GACCCCGTGAAGCAGCGGCGCGTGGTGTGGGCGTACGTCGGCGAGACCGACTCGGAGCGCGCCGACATCACCAAGGGGTG GGCCAACCTCCAGTCGATTCCGAGGACGGTGGAGCTTGACGAGAAGACCCGAACGAACCTCATCCAATGGCCCGTGGAGG AGCTCGATACCCTCCGCATCAACACCACCGATCTCAGCGGCTTCACCGTTGGCGCCGGCTCCGTCGTCCCCCTCCACCTC CACCAGACCGCTCAGCTTGACATCGAGGCAACCTTCCGCATCGATGCCTCCGCCATCGAGGCCCTCAACGAGGCCGATGT TAGCTACAACTGCACCACTAGCAGCGGCGCCGCCACCCGCGGCGCGCTTGGGCCCTTCGGCCTTCTTGTCCTCGCCAACC GCGCCCTGACCGAACAGACGGGAGTCTACTTCTATGTGTCCAAGGGCCTCGATGGTGGTCTTCGGACTCACTTCTGCCAC GACGAGTTGCGGTCGTCACACGCTAGTGATGTGGTGAAGCGGGTGGTGGGTAGCACGGTGCCGGTGCTCGACGGCGAGGA TTTCTCCGTCAGGGTGCTCGTGGACCACTCCATTGTCCAGAGCTTCGCGATGGGCGGGAGGTTGACGGCAACGTCGAGGG CATACCCGACCGAGGCCATCTACGCGGCAGCGGGGGTCTACATGTTCAACAACGCCACCGGCACTAGCGTCACCGCCGAG AAGCTTGTCGTGCATGATATGGACTCATCGTACAACCATATATACACAGATGATGACTTGGTAGTTGTCAATTAG |
Ta_4A:739415986-739415994:+ does not have available information here.