
| Microexon ID | Ta_2B:571516778-571516786:+ |
| Species | Triticum Aestivum | Coordinates | 2B:571516778..571516786 |
| Microexon Cluster ID | MEP22 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | Glyco_hydro_32N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | YSKYAMMGRACYGSYTWYCAYTTYCARCCYSMCAARAAYTGGATGAAYGATCCYAAYGGTCCAATGTWYTACAAGGGATKGTACCAYYTSTTCTAYCARTACAAYCCV |
| Logo of Microexon-tag DNA Seq | 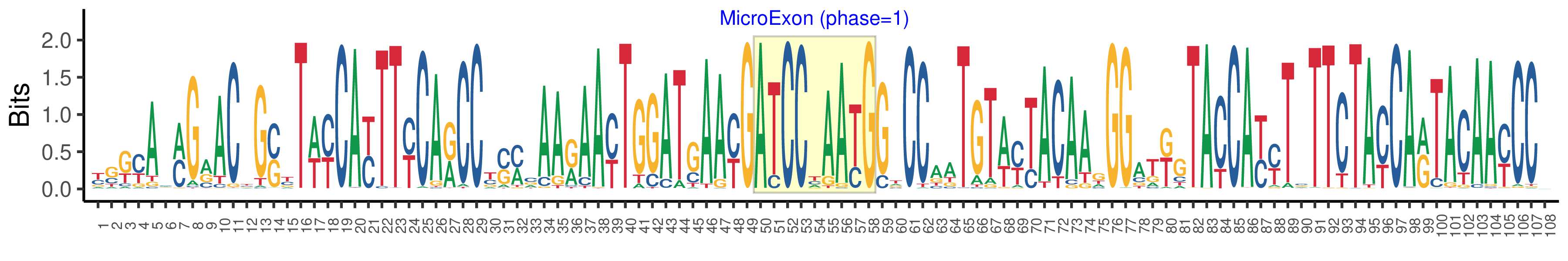 |
| Alignment of exons |  |
| Microexon DNA seq | ATCCCAACG |
| Microexon Amino Acid seq | DPNG |
| Microexon-tag DNA Seq | TGGCAGCGCACGGCGTTCCATTTCCAGCCCCAGAAGAATTGGATGAACGATCCCAACGGTCCACTGTATTACAAGGGATGGTATCACCTCTTCTACCAGTGGAACCCG |
| Microexon-tag Amino Acid Seq | WQRTAFHFQPQKNWMNDPNGPLYYKGWYHLFYQWNP |
| Microexon-tag spanning region | 571516566-571517029 |
| Microexon-tag prediction score | 0.9726 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | TraesCS2B02G403000.1x |
| Reference Transcript ID | TraesCS2B02G403000.1 |
| Gene ID | TraesCS2B02G403000 |
| Gene Name | NA |
| Transcript ID | TraesCS2B02G403000.1 |
| Protein ID | |
| Gene ID | TraesCS2B02G403000 |
| Gene Name | |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 4.50E-105 |
| Motif start | 125 |
| Motif end | 448 |
| Protein seq | >TraesCS2B02G403000.1 MIPAVPAIVDDAAQNANAPLLPETNLKGRRPPARQERPTTVLPAIAWTVLLLALVGLVVYPQGHGGGRADTIGDEVASAG RAVEVAASRGAVQGVSEKSTASALLGGGAYDWTNAMLAWQRTAFHFQPQKNWMNDPNGPLYYKGWYHLFYQWNPDGAVWG NITWGHAVSRDLVHWLHLPLAMVPDHWYDINGVWTGSATQLPDGRIVMLYTGSTEDGVQVQLLAEPADPSDPLLRRWAKS EANPVLVPPPGIGLTDFRDPTTAWLNPADKAWRITIGSKNQEHAGLALVYRTEDFLHYDLLPTLLHVVQGTGMWECVDFY PVSTEPAAGVGLDTSTASGAGVKHVLKTSLDDDRNDYYAIGTYDPATDRWAPDDAAIDVGIGLRYDYGKFYASKTFYDPV GRRRVLWGWIGESDSERADILKGWASLQSIPRTVLMDTKTGSNLLQWPVVEVENLRMRGKRFDGLDLQPGSVVPLGVGRA TQLDIEAVFQVQAGAAAAGGVAAGAEAPYNCSASAGAAGRGSLGPFGLLVLADDGLSEQTAVYFYLVKGADGKLSTHFCQ DAFRSSKANDLVKAVYGSLVPVLDGEDLSVRILVDHSIVESFAQGGRTCITSRVYPTKAIYDSARVFLFNNATNVNVTAK SIKIWELNSAYIRPYPDSS* |
| CDS seq | >TraesCS2B02G403000.1 ATGATCCCGGCCGTCCCGGCGATCGTGGACGACGCCGCCCAGAACGCGAACGCGCCGCTGCTTCCCGAGACCAACCTCAA GGGTCGCCGTCCTCCGGCACGGCAGGAGCGCCCGACGACCGTTCTCCCGGCCATCGCGTGGACCGTGCTCCTTCTCGCGC TCGTCGGGCTCGTCGTCTATCCACAAGGGCATGGCGGCGGCCGGGCCGACACGATAGGAGACGAGGTCGCGTCAGCGGGC CGCGCCGTGGAGGTGGCCGCCTCCCGCGGCGCCGTGCAAGGCGTGTCGGAGAAGTCCACCGCCTCGGCGCTCCTCGGCGG CGGCGCCTACGACTGGACCAACGCGATGCTGGCGTGGCAGCGCACGGCGTTCCATTTCCAGCCCCAGAAGAATTGGATGA ACGATCCCAACGGTCCACTGTATTACAAGGGATGGTATCACCTCTTCTACCAGTGGAACCCGGACGGGGCGGTGTGGGGG AACATCACGTGGGGCCACGCCGTGTCGCGCGACCTCGTCCACTGGCTCCACCTGCCGCTGGCCATGGTGCCCGACCACTG GTACGACATCAACGGCGTCTGGACCGGCTCGGCGACGCAGCTCCCCGACGGCAGGATCGTGATGCTCTACACCGGCTCCA CGGAGGACGGCGTGCAGGTGCAGCTCCTGGCCGAGCCGGCGGACCCGTCCGACCCGCTGCTGCGGCGCTGGGCCAAGTCG GAGGCGAACCCCGTGCTGGTGCCGCCGCCGGGCATCGGGCTCACGGACTTCCGCGACCCGACGACGGCGTGGCTAAACCC GGCCGACAAGGCCTGGCGGATCACCATCGGGTCCAAGAACCAGGAGCACGCCGGGCTGGCGCTGGTGTACAGGACCGAGG ACTTCCTGCACTACGACCTGCTGCCCACGCTGCTGCACGTCGTGCAGGGCACCGGGATGTGGGAGTGCGTGGACTTCTAC CCGGTGTCCACGGAGCCGGCCGCCGGCGTGGGGCTCGACACCTCCACCGCGTCGGGGGCCGGGGTGAAGCACGTGCTCAA GACCAGCCTCGACGACGACCGGAACGACTACTACGCGATCGGCACCTACGACCCCGCGACCGACCGGTGGGCGCCGGACG ACGCGGCGATCGACGTCGGGATCGGGCTCCGGTACGACTACGGCAAGTTCTACGCGTCCAAGACGTTCTACGACCCCGTG GGGCGGCGGCGCGTGCTGTGGGGGTGGATCGGCGAGTCCGACAGCGAGCGCGCCGACATCCTCAAGGGCTGGGCGTCCCT CCAGTCGATCCCGAGGACGGTTCTCATGGACACCAAGACGGGCAGCAACCTGCTCCAATGGCCGGTGGTGGAGGTGGAGA ACCTCCGAATGCGCGGCAAGCGCTTCGACGGCCTCGACCTGCAGCCCGGCTCCGTTGTGCCCCTGGGCGTCGGCAGAGCA ACACAGTTGGACATCGAGGCGGTCTTCCAGGTGCAGGCGGGCGCGGCGGCTGCCGGGGGCGTGGCGGCGGGGGCGGAAGC GCCGTACAACTGCAGCGCGAGCGCCGGCGCTGCGGGGCGGGGCTCGCTCGGCCCGTTCGGGCTGCTCGTGCTGGCCGACG ACGGCCTGTCGGAGCAGACCGCCGTCTACTTCTACCTGGTCAAGGGCGCGGACGGCAAGCTCAGCACCCACTTCTGCCAA GACGCCTTCAGGTCATCGAAGGCCAACGATCTCGTTAAGGCCGTCTACGGCAGCCTCGTCCCTGTGCTCGATGGAGAAGA TCTGTCAGTGAGAATACTGGTTGATCATTCCATCGTGGAGAGCTTTGCTCAGGGCGGGAGGACATGCATAACGTCGCGCG TGTACCCTACAAAGGCAATCTACGACTCTGCCAGAGTTTTCCTCTTCAACAACGCTACAAATGTTAACGTCACCGCAAAA TCGATCAAGATCTGGGAGCTCAATTCAGCCTACATCCGTCCATATCCAGATTCATCATAA |
Ta_2B:571516778-571516786:+ does not have available information here.