
| Microexon ID | Ha_13:114336722-114336730:- |
| Species | Helianthus annuus | Coordinates | 13:114336722..114336730 |
| Microexon Cluster ID | MEP22 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | Glyco_hydro_32N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | YSKYAMMGRACYGSYTWYCAYTTYCARCCYSMCAARAAYTGGATGAAYGATCCYAAYGGTCCAATGTWYTACAAGGGATKGTACCAYYTSTTCTAYCARTACAAYCCV |
| Logo of Microexon-tag DNA Seq | 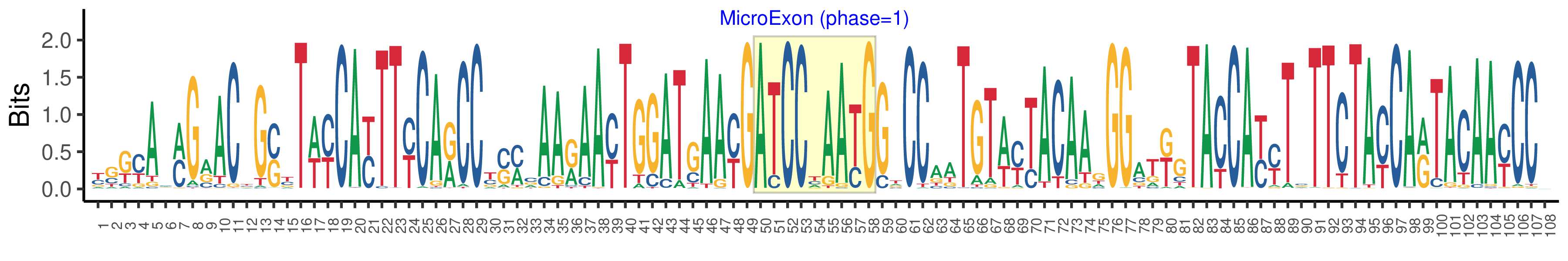 |
| Alignment of exons | 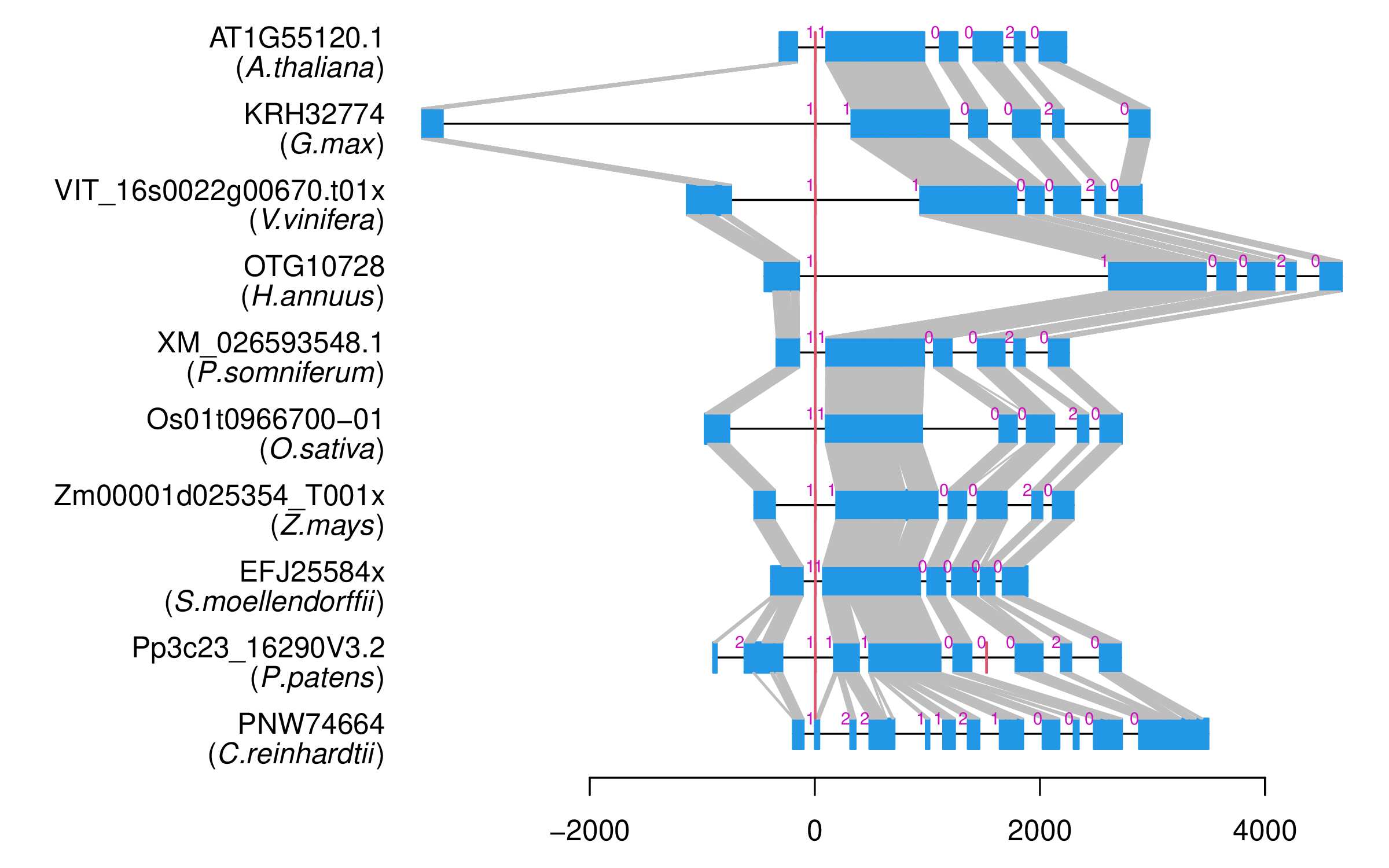 |
| Microexon DNA seq | ATCCTAATG |
| Microexon Amino Acid seq | DPNG |
| Microexon-tag DNA Seq | CCTTACAGGACAGCATATCATTTTCAGCCTTCCAAAAACTGGATGAATGATCCTAATGGACCAGTATATTACAAGGGGGTTTATCATTTGTTCTATCAATACAATCCA |
| Microexon-tag Amino Acid Seq | PYRTAYHFQPSKNWMNDPNGPVYYKGVYHLFYQYNP |
| Microexon-tag spanning region | 114336571-114337023 |
| Microexon-tag prediction score | 0.9526 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | OTG01908x |
| Reference Transcript ID | OTG01908 |
| Gene ID | HannXRQ_Chr13g0407161 |
| Gene Name | INV5 |
| Transcript ID | OTG01908 |
| Protein ID | OTG01908 |
| Gene ID | HannXRQ_Chr13g0407161 |
| Gene Name | INV5 |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 1.3e-102 |
| Motif start | 44 |
| Motif end | 359 |
| Protein seq | >OTG01908 MMKIYGFCVFSFCYFLISNGVQVAAFKDQRLRTNVSQPYRTAYHFQPSKNWMNDPNGPVYYKGVYHLFYQYNPYGPLWGN MSWGHAISYDLVNWVHLDIALLENEPYDINGCFSGSTTILQNGEPVIMYTGLDTDSHQVQNLAFPKNISDPLLSEWVKPS LNPVMSPPDRVDPTQFRDPTTGWLGADGEWRVVVGSELDGHGSATLYKSKDFRTWIKSPSPLHSSNKTIMWECPDFYPVL VSGKDGLDTSVQGKDVRHVLKASFNDHDYYILGNYDPTSDRYDVDVDFLDSKWSLRYDYGRFYASKSFYDSDKKRRILWS WVNEGDTDSDAFIKGWSGLQTIPRSIWLSENGDQLVQWPVEELEKLRTQKVHFENEELKGGSMIEISGITASQISHLACQ ISKMLSAEVVDPQALCKQKNASMSGKFGPFGLLVLASKDLTEYTAVFFRVFRTANNFRVLMCADQSRSSVRPEVDKSIYG AFLTLDPQYAKISLKTLIDHSIIESFGGEGLACITARVYPELAIGEQAHLYAFNNGTQSLSISSLSAWSMKNAQIVSTRK RRKPII* |
| CDS seq | >OTG01908 ATGATGAAGATTTATGGGTTTTGTGTATTTTCTTTTTGTTATTTCCTGATTTCTAATGGAGTCCAGGTTGCTGCTTTCAA GGATCAGAGATTGCGAACGAATGTTTCACAGCCTTACAGGACAGCATATCATTTTCAGCCTTCCAAAAACTGGATGAATG ATCCTAATGGACCAGTATATTACAAGGGGGTTTATCATTTGTTCTATCAATACAATCCATATGGTCCACTGTGGGGTAAC ATGTCATGGGGTCATGCTATATCATATGATCTTGTTAACTGGGTACATCTAGATATTGCTCTTTTAGAAAACGAGCCTTA CGATATCAATGGGTGCTTTTCTGGTTCGACAACGATTTTACAAAATGGGGAACCAGTAATCATGTACACAGGTCTTGATA CCGATTCACACCAAGTGCAAAACTTGGCATTTCCCAAAAACATATCGGATCCGCTACTAAGTGAGTGGGTAAAGCCGTCA CTTAACCCTGTAATGAGTCCTCCAGATAGAGTTGACCCAACACAGTTTAGAGATCCAACGACGGGTTGGTTGGGTGCAGA TGGAGAATGGAGGGTTGTGGTTGGAAGTGAGCTTGATGGTCATGGATCAGCAACTCTTTACAAAAGTAAGGATTTTCGTA CTTGGATTAAGTCCCCAAGCCCGTTACACTCTTCTAACAAGACAATAATGTGGGAATGTCCTGATTTTTATCCTGTTCTT GTTAGTGGAAAAGACGGGCTTGATACATCGGTTCAGGGGAAGGACGTTAGGCATGTTCTTAAAGCTAGTTTCAATGACCA TGATTATTATATACTGGGAAACTATGATCCGACAAGTGACCGGTATGACGTTGATGTTGACTTCTTGGACAGTAAATGGT CGTTACGGTATGATTATGGACGGTTTTATGCTTCGAAGTCATTTTATGATAGTGACAAGAAGAGAAGGATATTATGGTCA TGGGTTAATGAAGGTGATACTGATTCAGATGCTTTCATTAAAGGTTGGTCTGGCCTTCAGACGATTCCCAGGAGCATCTG GCTCAGTGAAAATGGAGATCAGTTGGTGCAGTGGCCCGTTGAGGAACTTGAGAAACTGCGAACACAGAAAGTGCATTTCG AGAATGAAGAACTCAAGGGCGGGTCCATGATTGAAATTTCTGGCATCACAGCTTCACAGATATCACATTTGGCTTGTCAA ATCTCGAAGATGCTGAGTGCTGAAGTAGTTGATCCCCAAGCTCTATGTAAACAAAAGAATGCTTCAATGAGTGGTAAATT CGGGCCTTTTGGTTTGCTGGTTTTGGCTTCGAAGGACCTGACTGAATACACTGCGGTCTTCTTTCGCGTTTTTAGAACTG CTAACAATTTCCGAGTGCTCATGTGTGCTGACCAAAGCAGGTCTTCTGTAAGACCAGAGGTTGACAAATCGATCTATGGA GCTTTTCTCACGCTTGATCCTCAATACGCAAAGATTTCCTTGAAAACCCTGATAGATCACTCCATTATAGAAAGTTTTGG TGGAGAAGGATTGGCTTGCATTACAGCAAGAGTTTATCCAGAATTGGCCATTGGAGAACAAGCACATCTTTATGCATTTA ACAATGGCACTCAGAGTTTGAGTATTTCAAGTTTAAGTGCTTGGAGTATGAAGAACGCTCAAATTGTTTCCACAAGGAAA AGAAGAAAGCCTATTATATAA |
| Microexon DNA seq | ATCCTAATG |
| Microexon Amino Acid seq | DPNG |
| Microexon-tag DNA Seq | CCTTACAGGACAGCATATCATTTTCAGCCTTCCAAAAACTGGATGAATGATCCTAATGGACCAGTATATTACAAGGGGGTTTATCATTTGTTCTATCAATACAATCCA |
| Microexon-tag Amino Acid seq | PYRTAYHFQPSKNWMNDPNGPVYYKGVYHLFYQYNP |
| Transcript ID | Ha.16356.1 |
| Gene ID | Ha.16356 |
| Gene Name | INV5 |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 5e-103 |
| Motif start | 44 |
| Motif end | 359 |
| Protein seq | >Ha.16356.1 MMKIYGFCVFSFCYFLISNGVQVAAFKDQRLRTNVSQPYRTAYHFQPSKNWMNDPNGPVYYKGVYHLFYQYNPYGPLWGN MSWGHAISYDLVNWVHLDIALLENEPYDINGCFSGSTTILQNGEPVIMYTGLDTDSHQVQNLAFPKNISDPLLSEWVKPS LNPVMSPPDRVDPTQFRDPTTGWLGADGEWRVVVGSELDGHGSATLYKSKDFRTWIKSPSPLHSSNKTIMWECPDFYPVL VSGKDGLDTSVQGKDVRHVLKASFNDHDYYILGNYDPTSDRYDVDVDFLDSKWSLRYDYGRFYASKSFYDSDKKRRILWS WVNEGDTDSDAFIKGWSGLQTIPRSIWLSENGDQLVQWPVEELEKLRTQKVHFENEELKGGSMIEISGITASQADVDITF GLSNLEDAEC* |
| CDS seq | >Ha.16356.1 ATGATGAAGATTTATGGGTTTTGTGTATTTTCTTTTTGTTATTTCCTGATTTCTAATGGAGTCCAGGTTGCTGCTTTCAA GGATCAGAGATTGCGAACGAATGTTTCACAGCCTTACAGGACAGCATATCATTTTCAGCCTTCCAAAAACTGGATGAATG ATCCTAATGGACCAGTATATTACAAGGGGGTTTATCATTTGTTCTATCAATACAATCCATATGGTCCACTGTGGGGTAAC ATGTCATGGGGTCATGCTATATCATATGATCTTGTTAACTGGGTACATCTAGATATTGCTCTTTTAGAAAACGAGCCTTA CGATATCAATGGGTGCTTTTCTGGTTCGACAACGATTTTACAAAATGGGGAACCAGTAATCATGTACACAGGTCTTGATA CCGATTCACACCAAGTGCAAAACTTGGCATTTCCCAAAAACATATCGGATCCGCTACTAAGTGAGTGGGTAAAGCCGTCA CTTAACCCTGTAATGAGTCCTCCAGATAGAGTTGACCCAACACAGTTTAGAGATCCAACGACGGGTTGGTTGGGTGCAGA TGGAGAATGGAGGGTTGTGGTTGGAAGTGAGCTTGATGGTCATGGATCAGCAACTCTTTACAAAAGTAAGGATTTTCGTA CTTGGATTAAGTCCCCAAGCCCGTTACACTCTTCTAACAAGACAATAATGTGGGAATGTCCTGATTTTTATCCTGTTCTT GTTAGTGGAAAAGACGGGCTTGATACATCGGTTCAGGGGAAGGACGTTAGGCATGTTCTTAAAGCTAGTTTCAATGACCA TGATTATTATATACTGGGAAACTATGATCCGACAAGTGACCGGTATGACGTTGATGTTGACTTCTTGGACAGTAAATGGT CGTTACGGTATGATTATGGACGGTTTTATGCTTCGAAGTCATTTTATGATAGTGACAAGAAGAGAAGGATATTATGGTCA TGGGTTAATGAAGGTGATACTGATTCAGATGCTTTCATTAAAGGTTGGTCTGGCCTTCAGACGATTCCCAGGAGCATCTG GCTCAGTGAAAATGGAGATCAGTTGGTGCAGTGGCCCGTTGAGGAACTTGAGAAACTGCGAACACAGAAAGTGCATTTCG AGAATGAAGAACTCAAGGGCGGGTCCATGATTGAAATTTCTGGCATCACAGCTTCACAGGCTGATGTAGATATCACATTT GGCTTGTCAAATCTCGAAGATGCTGAGTGCTGA |