
| Microexon ID | Gm_8:15445324-15445332:- |
| Species | Glycine max | Coordinates | 8:15445324..15445332 |
| Microexon Cluster ID | MEP22 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | Glyco_hydro_32N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | YSKYAMMGRACYGSYTWYCAYTTYCARCCYSMCAARAAYTGGATGAAYGATCCYAAYGGTCCAATGTWYTACAAGGGATKGTACCAYYTSTTCTAYCARTACAAYCCV |
| Logo of Microexon-tag DNA Seq | 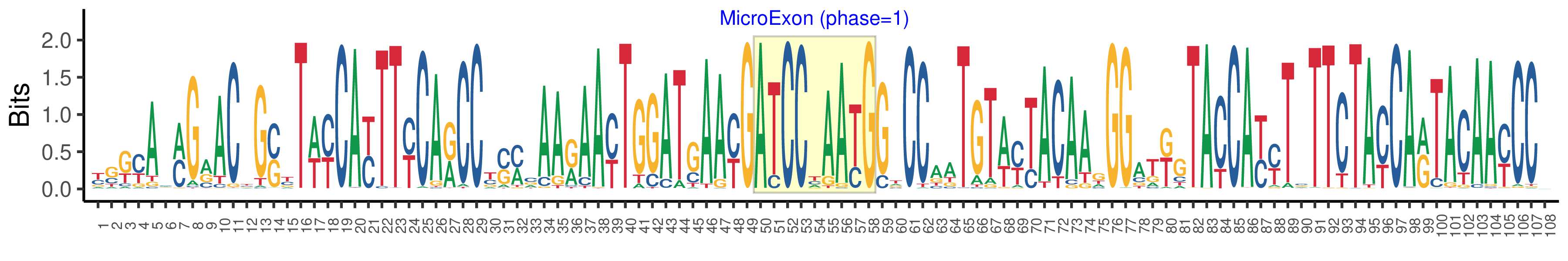 |
| Alignment of exons | 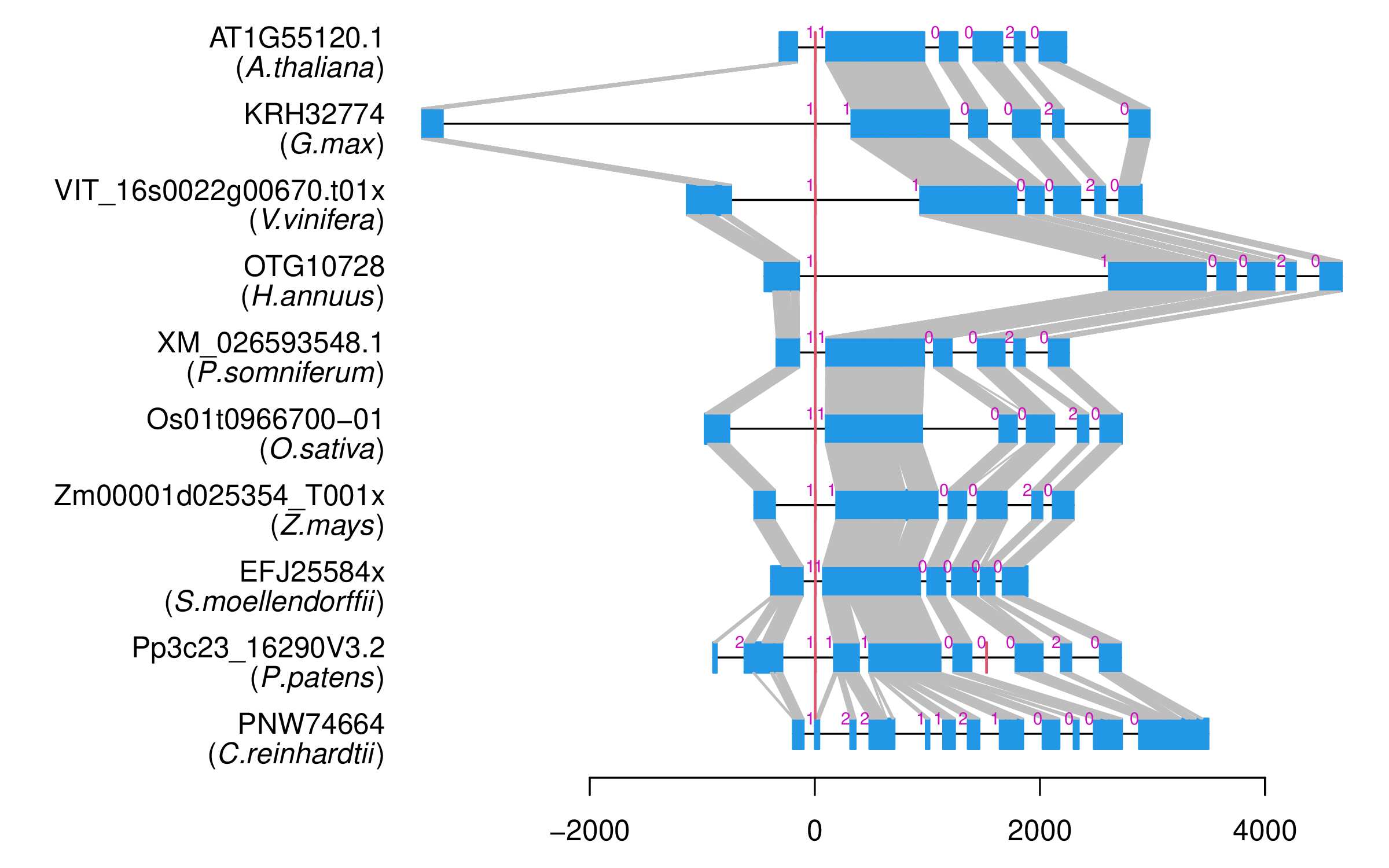 |
| Microexon DNA seq | ATCCCAATG |
| Microexon Amino Acid seq | DPNG |
| Microexon-tag DNA Seq | CCTTACAGAACTGCTTATCACTTCCAACCTCCCAATAATTGGATAAATGATCCCAATGGACCATTGAGATATGGAGGACTCTATCACCTGTTCTATCAATACAATCCT |
| Microexon-tag Amino Acid Seq | PYRTAYHFQPPNNWINDPNGPLRYGGLYHLFYQYNP |
| Microexon-tag spanning region | 15444567-15445554 |
| Microexon-tag prediction score | 0.9262 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH44130x |
| Reference Transcript ID | KRH44130 |
| Gene ID | GLYMA_08G192000 |
| Gene Name | NA |
| Transcript ID | KRH44130 |
| Protein ID | KRH44130 |
| Gene ID | GLYMA_08G192000 |
| Gene Name | NA |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 1.9e-107 |
| Motif start | 45 |
| Motif end | 362 |
| Protein seq | >KRH44130 MTMSTIWLLTLFSVIYGSAATHHIYRNLQSVSSDSSNQPYRTAYHFQPPNNWINDPNGPLRYGGLYHLFYQYNPKGAVWG NIVWAHSVSRDLVNWTPLDPAIFPSQSSDINGCWSGSATLLPGNKPAILYTGIDSLNQQVQNFAQPKNLSDPFLREWVKS PKNPLMAPTSANKINSSSFRDPTTAWLGKDGHWRVLVGSKRRTRGMAILYRSKDFVKWVQAKHPLHSTLGSGMWECPDFF PVLSNGQLGVDTSVNGEYVRHVLKVSLDDKKHDYYMIGSYNAAKDAFIPDEESNIFVLRYDYGKYYASKTFFDDGKKRRI LLGWVNESSSVADDIKKGWSGIHTIPRAIWLHKSGRQLVQWPVEEVEKLRAYPVNLLPQVLKGGKLLPINGVTASQADVE ISFEVSKLRKAEVLDYWTDPQILCSKKGSSVKSGLGPFGLLVFASEGLQEYTSVFFRIFRHQHKYLVLLCSDQNRSSLNK DNDLTSYGTFVDVDPLHDKLSLRTLIDHSVVESFGGEGRACITARVYPTLAINDKAQLYAFNNGTADVKITRLSAWSMKK AQIH* |
| CDS seq | >KRH44130 ATGACTATGTCTACTATTTGGCTTTTGACTCTCTTTTCTGTCATTTATGGCAGTGCAGCTACTCATCACATTTATAGAAA TCTTCAGAGTGTATCTTCTGATTCCTCCAACCAACCTTACAGAACTGCTTATCACTTCCAACCTCCCAATAATTGGATAA ATGATCCCAATGGACCATTGAGATATGGAGGACTCTATCACCTGTTCTATCAATACAATCCTAAAGGTGCAGTTTGGGGA AATATTGTGTGGGCACATTCAGTTTCAAGGGATCTTGTGAATTGGACCCCACTAGATCCTGCCATCTTTCCATCTCAATC GTCCGATATAAATGGTTGTTGGTCAGGATCAGCCACACTACTTCCCGGGAACAAACCAGCCATTTTGTATACTGGAATTG ACTCATTGAATCAGCAAGTTCAAAACTTTGCCCAACCAAAAAATTTGTCTGACCCATTTCTTAGGGAATGGGTTAAGTCC CCCAAAAATCCCCTAATGGCACCAACTAGTGCTAACAAGATCAATTCAAGTTCATTTAGGGACCCCACCACTGCTTGGCT AGGCAAAGATGGGCATTGGAGGGTACTTGTTGGAAGCAAAAGAAGAACTAGGGGAATGGCAATCTTGTATAGGAGCAAAG ACTTTGTTAAATGGGTTCAAGCCAAACACCCTTTGCATTCAACCCTAGGAAGTGGCATGTGGGAGTGTCCTGATTTTTTC CCTGTTTTGAGTAATGGCCAATTGGGTGTTGACACGTCGGTGAACGGTGAATATGTTAGGCACGTGCTCAAGGTTAGCTT AGATGACAAAAAACATGATTACTATATGATTGGGAGTTACAATGCTGCCAAGGATGCATTTATCCCAGATGAAGAGTCTA ATATATTTGTTTTAAGATATGATTATGGAAAATACTATGCCTCAAAAACTTTCTTTGATGATGGGAAGAAGAGAAGGATT TTGCTAGGGTGGGTCAATGAATCCTCTAGTGTTGCTGATGATATCAAGAAAGGATGGTCTGGAATCCATACAATTCCAAG GGCTATATGGCTTCATAAGTCTGGTAGACAGTTGGTACAGTGGCCAGTGGAAGAAGTTGAAAAGCTACGTGCATACCCAG TCAACTTGCTACCCCAAGTGCTAAAAGGAGGCAAGTTGCTTCCAATTAATGGTGTCACAGCATCACAGGCTGATGTTGAA ATTTCGTTTGAAGTGAGTAAGTTGAGAAAGGCTGAAGTATTGGACTATTGGACAGACCCCCAAATTCTATGTAGTAAAAA GGGTTCATCCGTCAAAAGTGGTTTGGGCCCATTTGGTCTACTAGTTTTTGCTTCAGAGGGTTTGCAAGAGTATACATCAG TTTTCTTCAGAATATTCAGACACCAACACAAATATTTGGTGCTCTTGTGCAGTGACCAAAACAGGTCTTCCTTAAACAAA GACAATGATTTAACCTCTTATGGAACTTTTGTGGACGTGGACCCTCTTCATGACAAGCTATCATTGAGAACCTTGATTGA TCATTCTGTCGTGGAGAGTTTTGGAGGAGAAGGAAGGGCTTGCATCACAGCAAGAGTTTATCCCACGTTGGCAATCAATG ATAAAGCACAACTATATGCTTTCAATAATGGAACTGCTGATGTCAAGATCACAAGATTGAGTGCTTGGAGCATGAAGAAA GCACAAATACATTGA |
| Microexon DNA seq | ATCCCAATG |
| Microexon Amino Acid seq | DPNG |
| Microexon-tag DNA Seq | CCTTACAGAACTGCTTATCACTTCCAACCTCCCAATAATTGGATAAATGATCCCAATGGACCATTGAGATATGGAGGACTCTATCACCTGTTCTATCAATACAATCCT |
| Microexon-tag Amino Acid seq | PYRTAYHFQPPNNWINDPNGPLRYGGLYHLFYQYNP |
| Transcript ID | KRH44130 |
| Gene ID | Gm.50068 |
| Gene Name | NA |
| Pfam domain motif | Glyco_hydro_32N |
| Motif E-value | 1.9e-107 |
| Motif start | 45 |
| Motif end | 362 |
| Protein seq | >KRH44130 MTMSTIWLLTLFSVIYGSAATHHIYRNLQSVSSDSSNQPYRTAYHFQPPNNWINDPNGPLRYGGLYHLFYQYNPKGAVWG NIVWAHSVSRDLVNWTPLDPAIFPSQSSDINGCWSGSATLLPGNKPAILYTGIDSLNQQVQNFAQPKNLSDPFLREWVKS PKNPLMAPTSANKINSSSFRDPTTAWLGKDGHWRVLVGSKRRTRGMAILYRSKDFVKWVQAKHPLHSTLGSGMWECPDFF PVLSNGQLGVDTSVNGEYVRHVLKVSLDDKKHDYYMIGSYNAAKDAFIPDEESNIFVLRYDYGKYYASKTFFDDGKKRRI LLGWVNESSSVADDIKKGWSGIHTIPRAIWLHKSGRQLVQWPVEEVEKLRAYPVNLLPQVLKGGKLLPINGVTASQADVE ISFEVSKLRKAEVLDYWTDPQILCSKKGSSVKSGLGPFGLLVFASEGLQEYTSVFFRIFRHQHKYLVLLCSDQNRSSLNK DNDLTSYGTFVDVDPLHDKLSLRTLIDHSVVESFGGEGRACITARVYPTLAINDKAQLYAFNNGTADVKITRLSAWSMKK AQIH* |
| CDS seq | >KRH44130 ATGACTATGTCTACTATTTGGCTTTTGACTCTCTTTTCTGTCATTTATGGCAGTGCAGCTACTCATCACATTTATAGAAA TCTTCAGAGTGTATCTTCTGATTCCTCCAACCAACCTTACAGAACTGCTTATCACTTCCAACCTCCCAATAATTGGATAA ATGATCCCAATGGACCATTGAGATATGGAGGACTCTATCACCTGTTCTATCAATACAATCCTAAAGGTGCAGTTTGGGGA AATATTGTGTGGGCACATTCAGTTTCAAGGGATCTTGTGAATTGGACCCCACTAGATCCTGCCATCTTTCCATCTCAATC GTCCGATATAAATGGTTGTTGGTCAGGATCAGCCACACTACTTCCCGGGAACAAACCAGCCATTTTGTATACTGGAATTG ACTCATTGAATCAGCAAGTTCAAAACTTTGCCCAACCAAAAAATTTGTCTGACCCATTTCTTAGGGAATGGGTTAAGTCC CCCAAAAATCCCCTAATGGCACCAACTAGTGCTAACAAGATCAATTCAAGTTCATTTAGGGACCCCACCACTGCTTGGCT AGGCAAAGATGGGCATTGGAGGGTACTTGTTGGAAGCAAAAGAAGAACTAGGGGAATGGCAATCTTGTATAGGAGCAAAG ACTTTGTTAAATGGGTTCAAGCCAAACACCCTTTGCATTCAACCCTAGGAAGTGGCATGTGGGAGTGTCCTGATTTTTTC CCTGTTTTGAGTAATGGCCAATTGGGTGTTGACACGTCGGTGAACGGTGAATATGTTAGGCACGTGCTCAAGGTTAGCTT AGATGACAAAAAACATGATTACTATATGATTGGGAGTTACAATGCTGCCAAGGATGCATTTATCCCAGATGAAGAGTCTA ATATATTTGTTTTAAGATATGATTATGGAAAATACTATGCCTCAAAAACTTTCTTTGATGATGGGAAGAAGAGAAGGATT TTGCTAGGGTGGGTCAATGAATCCTCTAGTGTTGCTGATGATATCAAGAAAGGATGGTCTGGAATCCATACAATTCCAAG GGCTATATGGCTTCATAAGTCTGGTAGACAGTTGGTACAGTGGCCAGTGGAAGAAGTTGAAAAGCTACGTGCATACCCAG TCAACTTGCTACCCCAAGTGCTAAAAGGAGGCAAGTTGCTTCCAATTAATGGTGTCACAGCATCACAGGCTGATGTTGAA ATTTCGTTTGAAGTGAGTAAGTTGAGAAAGGCTGAAGTATTGGACTATTGGACAGACCCCCAAATTCTATGTAGTAAAAA GGGTTCATCCGTCAAAAGTGGTTTGGGCCCATTTGGTCTACTAGTTTTTGCTTCAGAGGGTTTGCAAGAGTATACATCAG TTTTCTTCAGAATATTCAGACACCAACACAAATATTTGGTGCTCTTGTGCAGTGACCAAAACAGGTCTTCCTTAAACAAA GACAATGATTTAACCTCTTATGGAACTTTTGTGGACGTGGACCCTCTTCATGACAAGCTATCATTGAGAACCTTGATTGA TCATTCTGTCGTGGAGAGTTTTGGAGGAGAAGGAAGGGCTTGCATCACAGCAAGAGTTTATCCCACGTTGGCAATCAATG ATAAAGCACAACTATATGCTTTCAATAATGGAACTGCTGATGTCAAGATCACAAGATTGAGTGCTTGGAGCATGAAGAAA GCACAAATACATTGA |