
| Microexon ID | Zm_10:72230415-72230423:- |
| Species | Zea mays | Coordinates | 10:72230415..72230423 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 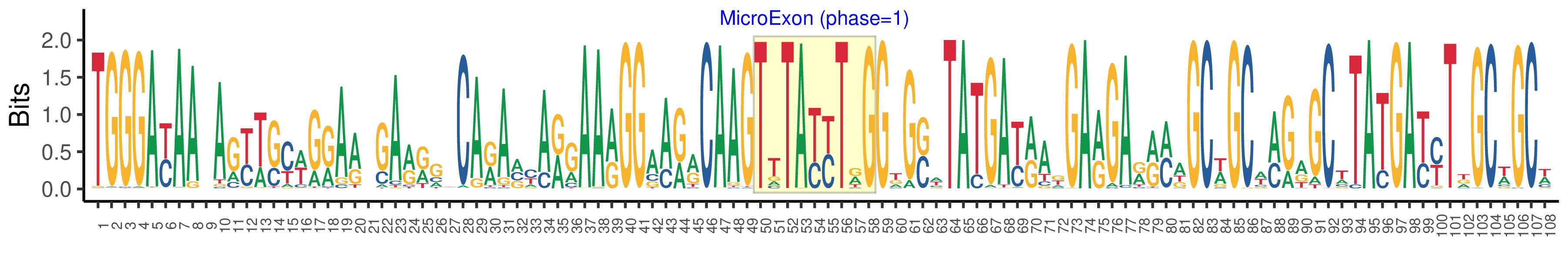 |
| Alignment of exons |  |
| Microexon DNA seq | TGTACTTAG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAACACCAGCCAGGTCGAAGGCCGCAAGCGCAAGGGCAAGCACGTGTACTTAGGCAGCTATGTCGATGAAGAGCAGGCTGCAAAGGCACATGATTTGGCTGCC |
| Microexon-tag Amino Acid Seq | WDNTSQVEGRKRKGKHVYLGSYVDEEQAAKAHDLAA |
| Microexon-tag spanning region | 72230285-72232021 |
| Microexon-tag prediction score | 0.9079 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Zm00001d024468_T001x |
| Reference Transcript ID | Zm00001d024468_T001 |
| Gene ID | Zm00001d024468 |
| Gene Name | AP2-EREBP-transcription factor 163 |
| Transcript ID | Zm00001d024468_T001 |
| Protein ID | Zm00001d024468_P001 |
| Gene ID | Zm00001d024468 |
| Gene Name | AP2-EREBP-transcription factor 163 |
| Pfam domain motif | AP2 |
| Motif E-value | 1.8e-09 |
| Motif start | 93 |
| Motif end | 151 |
| Protein seq | >Zm00001d024468_P001 MATVHPQSPAGPTHLPQEERHGDAPVPPAVSGEPAAAAEAGGEIAALDKQLAVVGGGEERRPAGAGAGAASAGGGGKLVA EAMRKYAAPRSSRFHGVTRLKWSGKYEAHLWDNTSQVEGRKRKGKHVYLGSYVDEEQAAKAHDLAALKYWGTGPNTKLNF NISDYEKEIEVMKTMSQDEFVAYIRRQSSCFSRETEEEAAEAYDIAAIELRGVHAVTNFDISNYCEDGLRKLEGPSEVAK LGSPSEVVKLAGQ* |
| CDS seq | >Zm00001d024468_T001 ATGGCCACCGTCCACCCTCAATCCCCGGCGGGTCCCACCCATCTCCCGCAGGAGGAGCGACACGGGGACGCCCCGGTACC GCCCGCCGTCTCCGGCGAACCCGCCGCCGCAGCGGAGGCGGGCGGCGAGATCGCCGCGCTCGACAAGCAGCTCGCCGTCG TCGGCGGCGGCGAGGAGAGGAGGCCGGCCGGGGCCGGGGCCGGGGCCGCCAGCGCAGGCGGGGGAGGGAAGCTGGTGGCG GAGGCCATGCGCAAGTACGCTGCCCCGCGGTCGTCGCGCTTCCACGGCGTCACCAGGCTCAAGTGGAGCGGCAAGTACGA GGCGCACCTTTGGGACAACACCAGCCAGGTCGAAGGCCGCAAGCGCAAGGGCAAGCACGTGTACTTAGGCAGCTATGTCG ATGAAGAGCAGGCTGCAAAGGCACATGATTTGGCTGCCCTTAAATATTGGGGCACAGGGCCAAACACCAAACTGAACTTC AATATTTCTGATTATGAGAAAGAAATTGAGGTTATGAAGACCATGTCCCAAGATGAATTTGTGGCCTACATAAGGAGACA AAGCAGTTGTTTCTCAAGAGAAACTGAAGAAGAGGCAGCAGAAGCTTATGACATTGCAGCGATCGAGCTTCGGGGTGTTC ATGCAGTCACCAACTTTGACATTAGTAATTACTGTGAAGATGGTTTGAGAAAATTAGAAGGCCCATCAGAGGTGGCTAAG CTTGGAAGTCCGTCGGAGGTTGTGAAGCTCGCTGGGCAATAG |
| Microexon DNA seq | TGTACTTAG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAACACCAGCCAGGTCGAAGGCCGCAAGCGCAAGGGCAAGCACGTGTACTTAGGCAGCTATGTCGATGAAGAGCAGGCTGCAAAGGCACATGATTTGGCTGCC |
| Microexon-tag Amino Acid seq | WDNTSQVEGRKRKGKHVYLGSYVDEEQAAKAHDLAA |
| Transcript ID | Zm00001d024468_T003 |
| Gene ID | Zm.6803 |
| Gene Name | AP2-EREBP-transcription factor 163 |
| Pfam domain motif | AP2 |
| Motif E-value | 2.2e-09 |
| Motif start | 93 |
| Motif end | 151 |
| Protein seq | >Zm00001d024468_T003 MATVHPQSPAGPTHLPQEERHGDAPVPPAVSGEPAAAAEAGGEIAALDKQLAVVGGGEERRPAGAGAGAASAGGGGKLVA EAMRKYAAPRSSRFHGVTRLKWSGKYEAHLWDNTSQVEGRKRKGKHVYLGSYVDEEQAAKAHDLAALKYWGTGPNTKLNF NISDYEKEIEVMKTMSQDEFVAYIRRQSSCFSRGTSSYRGVTRRKDGKWQARIGRIGESRDTKDIYLGTFETEEEAAEAY DIAAIELRGVHAVTNFDISNYCEDGLRKLEGPSEVAKLGSPSEVVKLAGQ* |
| CDS seq | >Zm00001d024468_T003 ATGGCCACCGTCCACCCTCAATCCCCGGCGGGTCCCACCCATCTCCCGCAGGAGGAGCGACACGGGGACGCCCCGGTACC GCCCGCCGTCTCCGGCGAACCCGCCGCCGCAGCGGAGGCGGGCGGCGAGATCGCCGCGCTCGACAAGCAGCTCGCCGTCG TCGGCGGCGGCGAGGAGAGGAGGCCGGCCGGGGCCGGGGCCGGGGCCGCCAGCGCAGGCGGGGGAGGGAAGCTGGTGGCG GAGGCCATGCGCAAGTACGCTGCCCCGCGGTCGTCGCGCTTCCACGGCGTCACCAGGCTCAAGTGGAGCGGCAAGTACGA GGCGCACCTTTGGGACAACACCAGCCAGGTCGAAGGCCGCAAGCGCAAGGGCAAGCACGTGTACTTAGGCAGCTATGTCG ATGAAGAGCAGGCTGCAAAGGCACATGATTTGGCTGCCCTTAAATATTGGGGCACAGGGCCAAACACCAAACTGAACTTC AATATTTCTGATTATGAGAAAGAAATTGAGGTTATGAAGACCATGTCCCAAGATGAATTTGTGGCCTACATAAGGAGACA AAGCAGTTGTTTCTCAAGAGGTACTTCATCATACCGGGGTGTAACCAGACGGAAGGATGGTAAATGGCAAGCAAGAATTG GTAGGATCGGTGAGAGTAGAGACACCAAAGACATTTACCTTGGGACCTTTGAAACTGAAGAAGAGGCAGCAGAAGCTTAT GACATTGCAGCGATCGAGCTTCGGGGTGTTCATGCAGTCACCAACTTTGACATTAGTAATTACTGTGAAGATGGTTTGAG AAAATTAGAAGGCCCATCAGAGGTGGCTAAGCTTGGAAGTCCGTCGGAGGTTGTGAAGCTCGCTGGGCAATAG |