
| Microexon ID | Zm_1:51467293-51467301:- |
| Species | Zea mays | Coordinates | 1:51467293..51467301 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 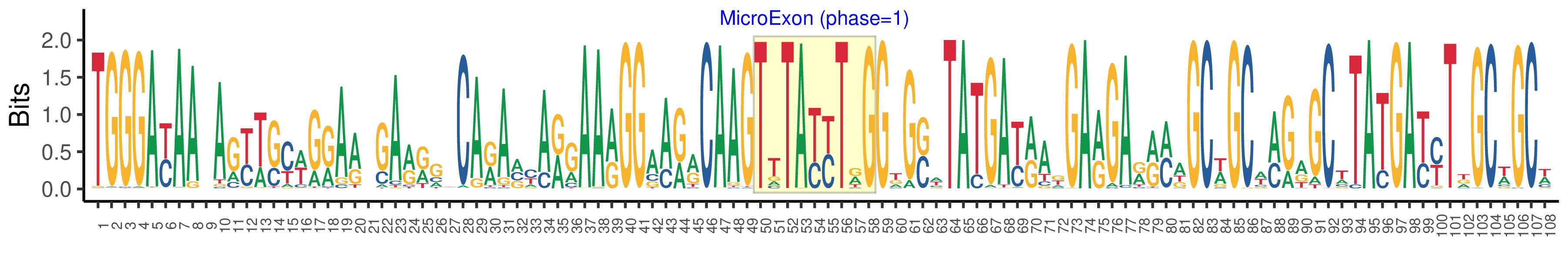 |
| Alignment of exons | 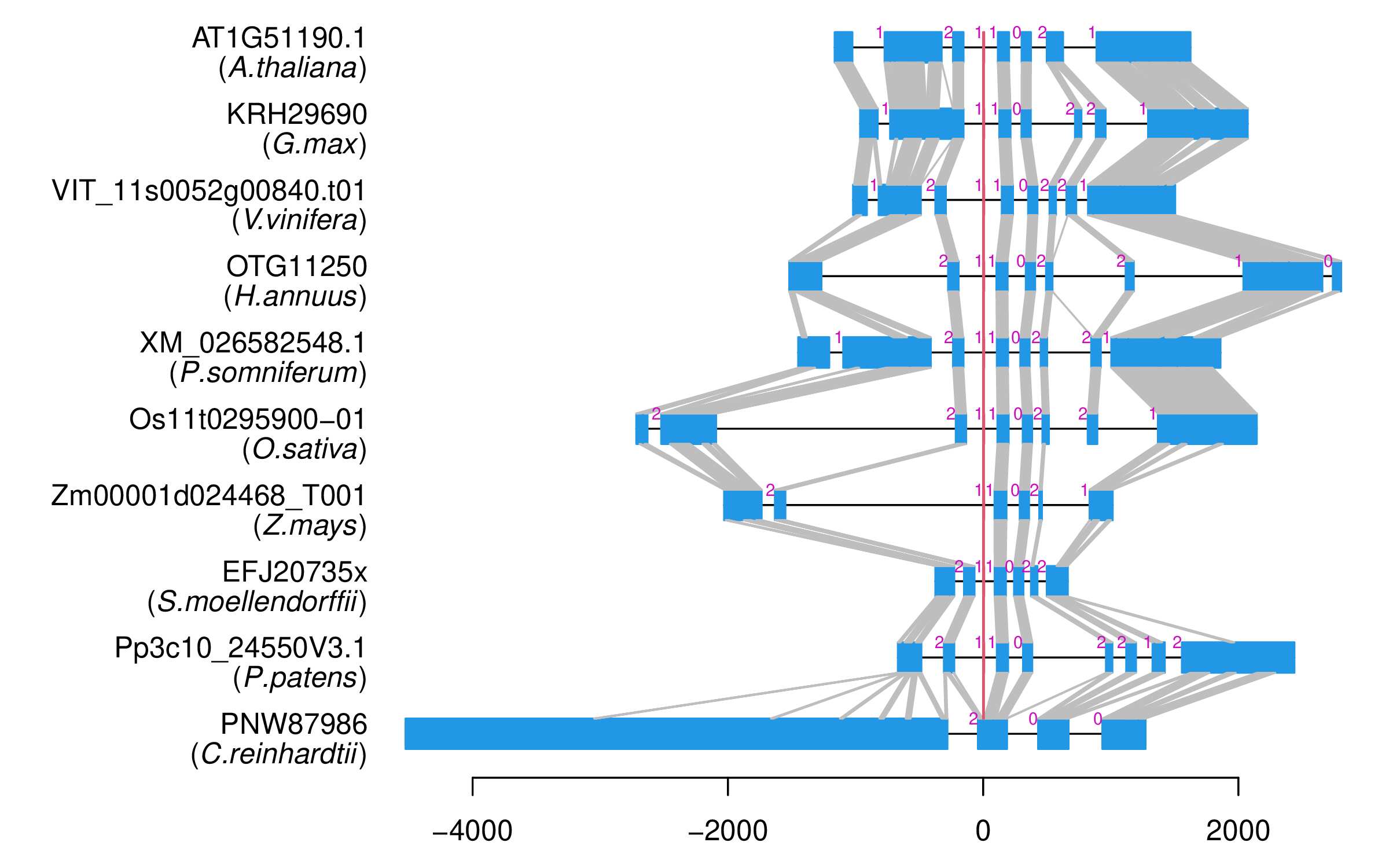 |
| Microexon DNA seq | TCTATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAACACGTGCCGTAAGGAAGGCCAGAAGCGCAAGGGGCGCCAAGTCTATTTGGGTGGCTACGACAAAGAAGATAAGGCTGCAAGGGCATATGACATTGCCGCT |
| Microexon-tag Amino Acid Seq | WDNTCRKEGQKRKGRQVYLGGYDKEDKAARAYDIAA |
| Microexon-tag spanning region | 51467160-51470195 |
| Microexon-tag prediction score | 0.9376 |
| Overlapped with the annotated transcript (%) | 91.67 |
| New Transcript ID | Zm00001d028919_T001x |
| Reference Transcript ID | Zm00001d028919_T001 |
| Gene ID | Zm00001d028919 |
| Gene Name | AP2-EREBP-transcription factor 26 |
Zm_1:51467293-51467301:- does not have available information here.
| Microexon DNA seq | TCTATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAACACGTGCCGTAAGGAAGGCCAGAAGCGCAAGGGGCGCCAAGTCTATTTGGGTGGCTACGACAAAGAAGATAAGGCTGCAAGGGCATATGACATTGCCGCT |
| Microexon-tag Amino Acid seq | WDNTCRKEGQKRKGRQVYLGGYDKEDKAARAYDIAA |
| Transcript ID | Zm.1313.1 |
| Gene ID | Zm.1313 |
| Gene Name | AP2-EREBP-transcription factor 26 |
| Pfam domain motif | AP2 |
| Motif E-value | 1.7e-11 |
| Motif start | 148 |
| Motif end | 206 |
| Protein seq | >Zm.1313.1 MSPPTNGAISLAYAPSMMLGAGALTNPPLLPFDGFTDEDFLASADAALLGEAGTDQTLLLLPSCPGANCCGGSSSDQGLG ALACEVTTAGSFSLLGQPAPGQVSWEVTTAVAADRNTFSRARDPAPSPPPSPALPLVQTTSQSQRTSIYRGVTRHRWTGR YEAHLWDNTCRKEGQKRKGRQVYLGGYDKEDKAARAYDIAALKYWGDNATTNFPRENYIREIQDMQNMNRRDVVASLRRK SSGFSRGASIYRGVTKHHQHGRWQARIGRVAGNKDLYLGTFATEQEAAEAYDIAALKFRGENAVTNFEPSRYNLLAIAQR DIPILGRKLIQKPAPEAEDQAALSARSFSQSQQSSNSLPPYFLTNLLQPLPSQHSLAQALPSYNNLGFGEPSLYWPCPCG DPGEQKVQLGSKLEIVDGLVQLANSAAN* |
| CDS seq | >Zm.1313.1 ATGTCGCCACCGACGAACGGCGCCATCTCGCTCGCCTACGCGCCGTCCATGATGCTGGGGGCGGGCGCCCTGACGAACCC GCCGCTCCTCCCCTTCGACGGCTTCACGGATGAGGACTTCCTCGCCTCCGCCGACGCCGCTCTCCTTGGCGAGGCTGGTA CCGACCAGACCCTCCTCCTGCTGCCCTCGTGCCCCGGGGCGAACTGCTGCGGCGGCAGTAGCTCCGACCAGGGTCTCGGA GCCCTAGCGTGTGAGGTCACAACCGCCGGATCCTTCTCGCTCTTGGGCCAGCCTGCGCCTGGGCAGGTTTCGTGGGAGGT GACGACTGCCGTCGCTGCGGACCGCAACACCTTCTCGCGCGCACGCGACCCTGCGCCCTCACCACCACCGTCCCCGGCGC TGCCGCTCGTGCAAACCACCTCCCAGTCCCAGAGGACCTCCATCTACCGCGGGGTGACCAGGCACAGGTGGACGGGGCGG TACGAGGCGCACCTGTGGGACAACACGTGCCGTAAGGAAGGCCAGAAGCGCAAGGGGCGCCAAGTCTATTTGGGTGGCTA CGACAAAGAAGATAAGGCTGCAAGGGCATATGACATTGCCGCTCTGAAGTATTGGGGTGATAATGCTACTACTAACTTCC CTAGAGAGAACTACATCAGAGAGATACAGGATATGCAGAACATGAACAGGCGTGATGTTGTAGCATCACTGAGGAGGAAG AGCAGTGGCTTTTCTAGAGGAGCATCAATCTACAGAGGAGTAACAAAGCACCACCAACATGGACGCTGGCAGGCAAGAAT CGGCCGTGTTGCAGGGAACAAGGATCTGTATCTTGGAACTTTCGCAACCGAGCAAGAGGCTGCAGAGGCGTACGACATCG CGGCACTGAAGTTCAGGGGCGAGAATGCCGTGACCAACTTCGAGCCCAGCCGGTACAACCTGCTAGCGATCGCTCAACGA GACATCCCCATCCTGGGGAGGAAGCTGATTCAGAAGCCAGCTCCGGAGGCAGAAGACCAGGCCGCTCTGAGCGCTCGTTC GTTCTCCCAGTCCCAGCAGTCGAGCAACAGCTTGCCACCGTATTTCCTCACCAACCTCCTGCAGCCGCTGCCGTCGCAGC ACTCACTAGCTCAGGCTCTGCCGAGCTACAACAACTTGGGGTTCGGGGAGCCCAGCTTGTACTGGCCCTGCCCCTGCGGT GACCCGGGGGAGCAGAAGGTGCAGCTTGGCAGCAAGCTGGAAATCGTGGACGGTCTCGTGCAGCTGGCCAACTCTGCTGC GAACTGA |