
| Microexon ID | Zm_1:214379994-214380002:- |
| Species | Zea mays | Coordinates | 1:214379994..214380002 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 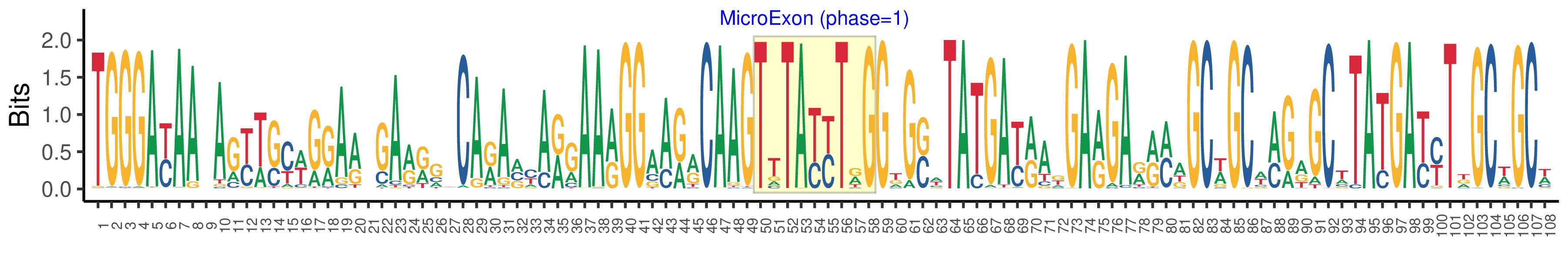 |
| Alignment of exons |  |
| Microexon DNA seq | TTTATCTCG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAAGGACGCCCGCAACGGCTCCCGGAGCAAGAAAGGCAAGCAAGTTTATCTCGGCGCGTACGACGACGAGGACGCGGCGGCGCGCGCGCACGACCTGGCGGCG |
| Microexon-tag Amino Acid Seq | WDKDARNGSRSKKGKQVYLGAYDDEDAAARAHDLAA |
| Microexon-tag spanning region | 214379786-214380241 |
| Microexon-tag prediction score | 0.8975 |
| Overlapped with the annotated transcript (%) | 91.67 |
| New Transcript ID | Zm00001d032144_T001x |
| Reference Transcript ID | Zm00001d032144_T001 |
| Gene ID | Zm00001d032144 |
| Gene Name | AP2-EREBP-transcription factor 60 |
Zm_1:214379994-214380002:- does not have available information here.
| Microexon DNA seq | TTTATCTCG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAAGGACGCCCGCAACGGCTCCCGGAGCAAGAAAGGCAAGCAAGTTTATCTCGGCGCGTACGACGACGAGGACGCGGCGGCGCGCGCGCACGACCTGGCGGCG |
| Microexon-tag Amino Acid seq | WDKDARNGSRSKKGKQVYLGAYDDEDAAARAHDLAA |
| Transcript ID | Zm.3645.2 |
| Gene ID | Zm.3645 |
| Gene Name | AP2-EREBP-transcription factor 60 |
| Pfam domain motif | AP2 |
| Motif E-value | 3.1e-12 |
| Motif start | 45 |
| Motif end | 103 |
| Protein seq | >Zm.3645.2 MARPRKNGGTDEDDANAATGATGKPKKLMKRARRKSESPSPRSSAYRGVTRHRWTGRFEAHLWDKDARNGSRSKKGKQVY LGAYDDEDAAARAHDLAALKYWGPAGTVLNFPLSGYDEERREMEGQPREEYVASLRRRSSGFARGVSKYRGVARHHHNGR WEARIGRVLGNKYLYLGTYATQEEAAVAYDMAAIEHRGFNAVTNFDISHYINHWHRHCHGPCDGSLGAMDVAPNVSLELD LLECPATVGLGLEETTGDDEFHNREDYLGHLFGVQQLPDEMGPPAHQMAPASSALDLVLQSPRFKELMQQVSAAGASETN GGSMRSSPSTSLCSFSPSPLELPSPPLQQPTEFIDGAPPRCSFPDDVQSFFDFKNDNDMSFVYAEVDTFLFGDLGAYAPP MFDFDLYE* |
| CDS seq | >Zm.3645.2 ATGGCCAGGCCCCGCAAGAACGGCGGTACCGACGAGGATGATGCTAACGCCGCCACGGGAGCCACCGGGAAGCCGAAGAA GCTGATGAAGCGGGCGCGGAGGAAGAGCGAGAGCCCCTCCCCGCGCAGCTCCGCGTACCGCGGCGTCACACGGCACCGGT GGACGGGGCGGTTCGAGGCGCACCTGTGGGACAAGGACGCCCGCAACGGCTCCCGGAGCAAGAAAGGCAAGCAAGTTTAT CTCGGCGCGTACGACGACGAGGACGCGGCGGCGCGCGCGCACGACCTGGCGGCGCTCAAGTACTGGGGCCCCGCCGGCAC CGTCCTCAACTTCCCGCTGAGCGGCTACGACGAGGAGCGGAGGGAGATGGAGGGGCAGCCCCGGGAGGAGTACGTCGCGT CGCTGCGGCGGCGGAGCAGCGGCTTCGCGCGAGGGGTGTCCAAGTACAGGGGCGTTGCCAGGCATCATCACAACGGGAGA TGGGAGGCGAGGATCGGGCGCGTGTTAGGCAACAAGTACCTCTATCTCGGCACTTACGCAACACAAGAGGAGGCGGCAGT GGCGTATGACATGGCGGCTATAGAACACCGCGGCTTCAACGCGGTCACCAACTTCGACATCAGCCATTACATCAACCACT GGCATCGACATTGCCATGGCCCCTGCGACGGCAGCCTCGGCGCGATGGACGTCGCACCGAACGTCTCGCTCGAACTAGAC CTACTCGAGTGCCCGGCCACCGTCGGCCTCGGCCTCGAGGAGACCACGGGCGACGATGAATTCCACAACCGCGAGGACTA CCTGGGGCACCTTTTCGGCGTGCAGCAGCTTCCCGACGAGATGGGGCCGCCGGCTCATCAGATGGCACCAGCATCGTCGG CGCTCGATCTGGTGTTGCAGTCGCCTAGGTTCAAGGAGCTGATGCAACAGGTTTCCGCGGCGGGAGCCTCCGAGACCAAC GGCGGCAGCATGCGTAGCTCGCCATCAACGTCCCTGTGTTCGTTCTCGCCTTCGCCTCTGGAGCTGCCATCGCCGCCGCT ACAGCAACCTACTGAATTCATCGATGGCGCGCCGCCGCGGTGCAGCTTCCCGGATGACGTGCAGTCGTTCTTCGATTTCA AGAACGATAACGACATGAGCTTCGTCTACGCGGAGGTTGACACATTCCTCTTCGGCGACCTAGGCGCATACGCTCCGCCA ATGTTCGACTTCGACCTGTACGAGTGA |