
| Microexon ID | Zm_1:16358348-16358356:- |
| Species | Zea mays | Coordinates | 1:16358348..16358356 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 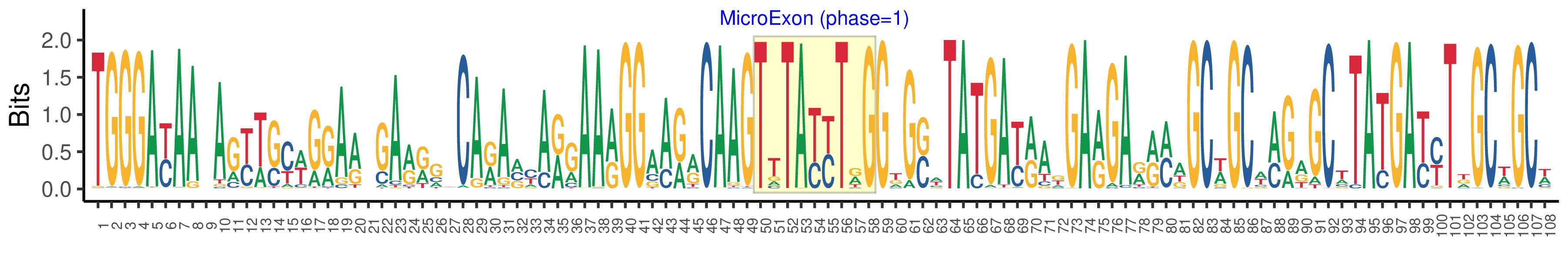 |
| Alignment of exons |  |
| Microexon DNA seq | TCTACCTAG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAACACTTGCAGGAAGGAAGGCCAGACTAGGAAAGGGAGGCAAGTCTACCTAGGCGGCTATGACAGGGAGGAGAAGGCTGCGAGGGCTTATGACCTGGCAGCT |
| Microexon-tag Amino Acid Seq | WDNTCRKEGQTRKGRQVYLGGYDREEKAARAYDLAA |
| Microexon-tag spanning region | 16358214-16358534 |
| Microexon-tag prediction score | 0.9715 |
| Overlapped with the annotated transcript (%) | 91.67 |
| New Transcript ID | Zm00001d027878_T002x |
| Reference Transcript ID | Zm00001d027878_T002 |
| Gene ID | Zm00001d027878 |
| Gene Name | AP2-EREBP-transcription factor 130 |
Zm_1:16358348-16358356:- does not have available information here.
| Microexon DNA seq | TCTACCTAG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAACACTTGCAGGAAGGAAGGCCAGACTAGGAAAGGGAGGCAAGTCTACCTAGGCGGCTATGACAGGGAGGAGAAGGCTGCGAGGGCTTATGACCTGGCAGCT |
| Microexon-tag Amino Acid seq | WDNTCRKEGQTRKGRQVYLGGYDREEKAARAYDLAA |
| Transcript ID | Zm.507.1 |
| Gene ID | Zm.507 |
| Gene Name | AP2-EREBP-transcription factor 130 |
| Pfam domain motif | AP2 |
| Motif E-value | 2.6e-12 |
| Motif start | 271 |
| Motif end | 329 |
| Protein seq | >Zm.507.1 MDMNNGWLGFSLSPSAASRGGYGYGDGGGGASASACGDGEGSCPSPAAAASPLPLVAMPLDDSLHYSSAPDWRHGAAEAK GPKLEDFMSITCSNKSSGRSLYDSCGHHDDEQASKYHEVHGIHPLSCGSYYHGCISSGGGGGGGIGLGINMNAPPCTGGF PDHQHHQFVPSSHHGQYFLGAPAASAGPPAGAAMPMYNAGGGSVVGGSMSISGIKSWLREAMYVPPERPAAAALSLAVTD DVPPAEPPQLLPAPLPVHRKPAQTFGQRTSQFRGVTRHRWTGRYEAHLWDNTCRKEGQTRKGRQVYLGGYDREEKAARAY DLAALKYWGPSTHINFPLSHYEKELEEMKHMSRQEFIAHLRRNSSGFSRGASMYRGVTRHHQHGRWQARIGRVAGNKDLY LGTFSTQEEAAEAYDIAAIKFRGLNAVTNFDISKYDVKRICASTHLIGGGDACRRSPTRPPDAAPALAGGADRSSDAPGD QAASDNSDTSDGHRGAHLLHGLQYGHPMKLEAGEGSSWMAAAAAARPVPGVHQLPMFALWNDC* |
| CDS seq | >Zm.507.1 ATGGACATGAACAACGGGTGGCTTGGCTTCTCCTTGTCTCCTTCGGCAGCGAGCAGGGGCGGCTACGGCTACGGCGACGG AGGCGGCGGAGCGAGCGCGAGCGCGTGCGGGGATGGAGAGGGCTCTTGCCCCTCGCCTGCCGCGGCTGCCTCGCCTCTCC CTCTCGTGGCCATGCCGCTCGATGACTCTCTGCACTATAGCTCTGCTCCAGATTGGAGGCACGGCGCAGCAGAAGCCAAA GGGCCCAAGCTCGAGGACTTCATGAGCATCACTTGCAGCAACAAGAGCAGTGGCCGCAGCCTCTACGACAGCTGCGGCCA CCACGACGACGAGCAGGCCAGCAAGTACCACGAGGTCCACGGCATCCACCCCCTCTCGTGCGGGTCGTACTACCACGGCT GCATCAGCAGCGGCGGCGGCGGCGGAGGCGGCATTGGCCTGGGGATCAACATGAACGCGCCGCCCTGCACCGGCGGGTTC CCGGACCACCAGCACCACCAGTTCGTGCCGTCGTCGCACCACGGCCAGTACTTCCTGGGCGCCCCAGCCGCAAGCGCGGG CCCGCCAGCAGGCGCAGCAATGCCGATGTACAACGCCGGCGGCGGCAGCGTTGTCGGCGGCTCCATGTCCATCTCCGGCA TCAAGTCCTGGCTCCGGGAGGCCATGTACGTGCCTCCGGAGCGACCGGCGGCGGCTGCGCTGTCGCTCGCCGTGACGGAT GACGTCCCGCCGGCCGAGCCGCCGCAGCTGCTGCCGGCGCCGTTGCCGGTGCACCGCAAGCCCGCGCAGACGTTCGGGCA GCGGACGTCGCAGTTCCGTGGCGTCACCAGGCACAGATGGACCGGGCGGTACGAGGCTCACCTGTGGGACAACACTTGCA GGAAGGAAGGCCAGACTAGGAAAGGGAGGCAAGTCTACCTAGGCGGCTATGACAGGGAGGAGAAGGCTGCGAGGGCTTAT GACCTGGCAGCTCTCAAGTACTGGGGCCCCTCGACTCACATAAATTTCCCGCTGAGCCACTACGAGAAGGAGCTGGAGGA GATGAAGCACATGTCGAGGCAGGAGTTCATTGCACACCTGAGAAGGAACAGCAGCGGCTTCTCGAGGGGAGCATCCATGT ACAGAGGAGTGACAAGGCATCACCAGCACGGGAGATGGCAGGCCAGGATTGGGAGGGTCGCTGGCAATAAGGACCTGTAC CTTGGCACATTCAGCACGCAGGAGGAAGCGGCGGAGGCGTACGACATCGCGGCGATCAAGTTCCGTGGCCTCAACGCGGT GACCAACTTCGACATCAGCAAGTACGACGTGAAGCGCATCTGCGCCAGCACCCACCTCATCGGCGGCGGCGACGCGTGCA GGCGCTCCCCGACGCGGCCGCCGGACGCGGCGCCGGCGCTCGCAGGTGGCGCCGACCGCTCGTCGGACGCGCCGGGCGAC CAAGCCGCGTCCGACAACTCGGACACCTCCGACGGCCACCGCGGCGCCCACCTGCTGCACGGCCTCCAGTACGGCCACCC CATGAAACTGGAGGCCGGGGAGGGAAGCAGCTGGATGGCGGCGGCGGCGGCAGCGCGGCCGGTGCCCGGCGTGCATCAGC TACCGATGTTCGCGCTGTGGAACGACTGTTGA |