
| Microexon ID | Ps_NC_039367.1:17897353-17897361:- |
| Species | Papaver somniferum | Coordinates | NC_039367.1:17897353..17897361 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 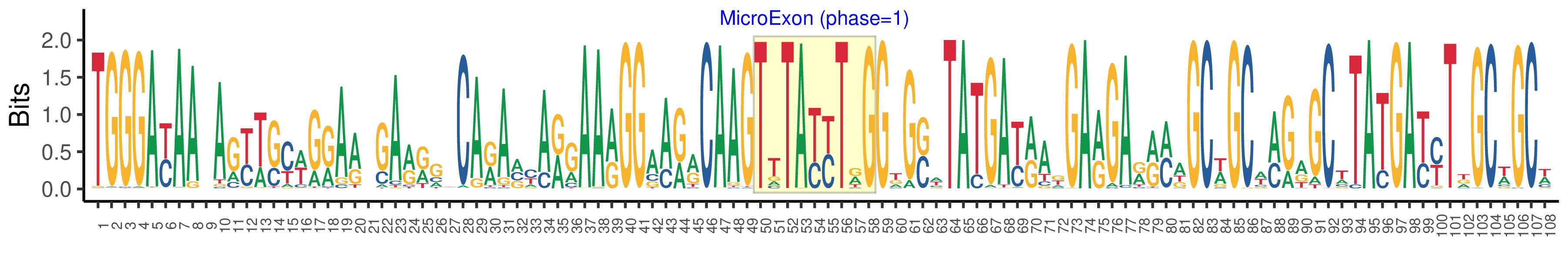 |
| Alignment of exons | 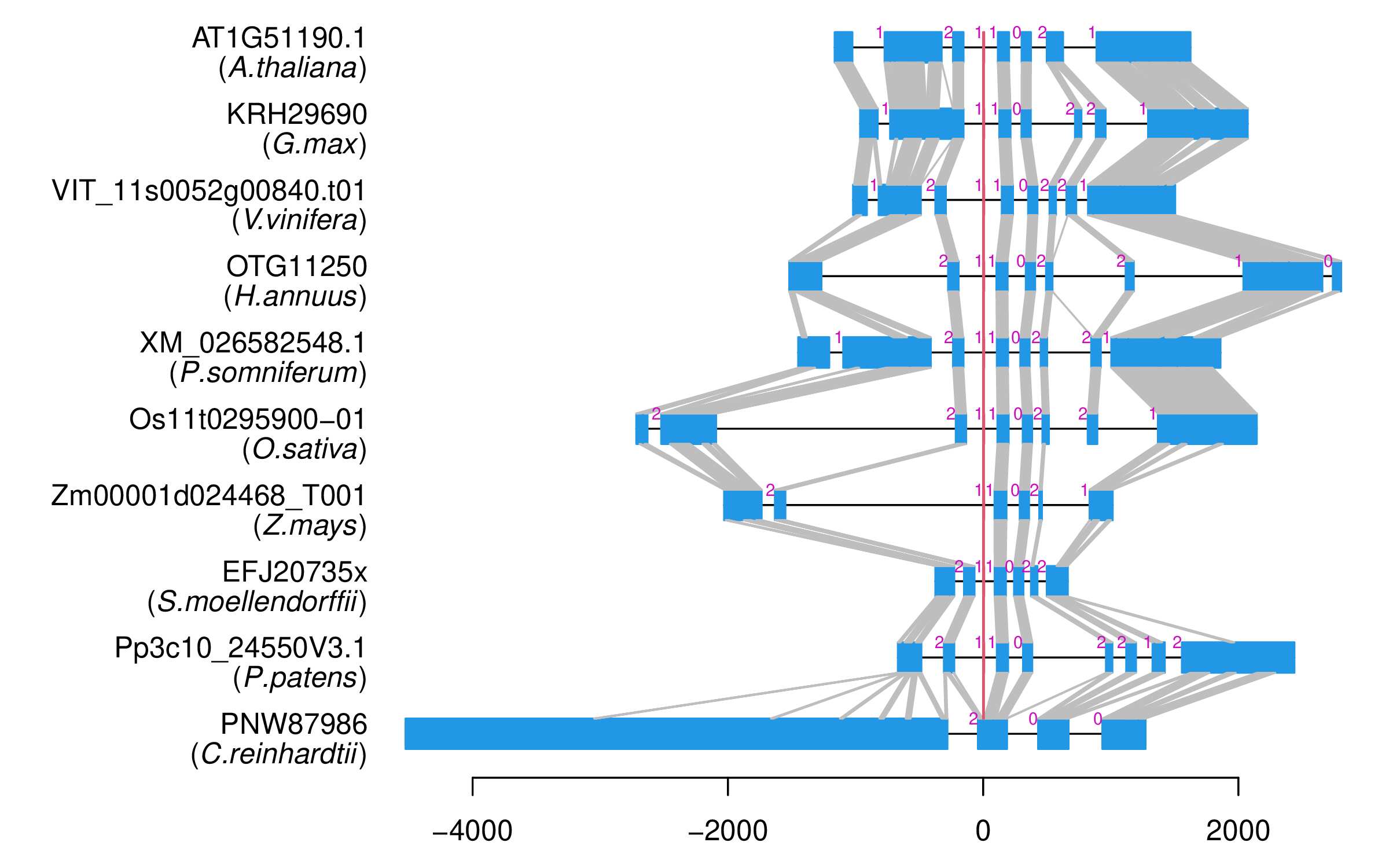 |
| Microexon DNA seq | TATATCTTG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAACACTACCATAAGTGAAACCCGGAAACGCAAAGGAAAGCAAGTATATCTTGGTGGATATGAAACTGAAGAGAAAGCAGCACAAGCATATGACCTTGCAGCT |
| Microexon-tag Amino Acid Seq | WDNTTISETRKRKGKQVYLGGYETEEKAAQAYDLAA |
| Microexon-tag spanning region | 17897208-17898019 |
| Microexon-tag prediction score | 0.9423 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026566716.1x |
| Reference Transcript ID | XM_026566716.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026566716.1 |
| Protein ID | XP_026422501.1 |
| Gene ID | LOC113318548 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 4.9e-11 |
| Motif start | 82 |
| Motif end | 144 |
| Protein seq | >XP_026422501.1 MGTLIPASENEICLLNQEVSLIDQVSIVADYERLPSSNDSSSKLIPPPSSSSSVNVNPASSSATGNEQVKKALYKVGQRT SNFRGVTSLFCRHRWTGKFEAHLWDNTTISETRKRKGKQVYLGGYETEEKAAQAYDLAALKLWGAGAHSKMNFPIPNYAK EVEEMKDMSREEYINSLRRKSICFARGNSIYRGVTRQHEGRRWQARIGRVAGGRDIYLGTYDTEEEAAEAYDIAAVKLRG MSAITNFHISNYYKEGSKELDIVPIDVTEKELPKVKIRRFIMP* |
| CDS seq | >XM_026566716.1 ATGGGAACTCTGATTCCTGCTTCAGAAAATGAGATATGTTTATTAAACCAGGAGGTATCCCTTATAGACCAAGTTTCAAT AGTGGCAGACTATGAGAGATTACCTTCTTCCAATGATTCATCTTCTAAATTAATTCCACCACCATCAAGTAGCAGTAGTG TTAATGTAAATCCTGCAAGCAGTAGTGCAACTGGAAACGAACAAGTAAAGAAGGCATTATATAAAGTTGGTCAGAGAACA TCGAATTTTCGTGGAGTTACTAGTTTGTTCTGCAGACACAGATGGACTGGCAAATTTGAAGCACATCTTTGGGATAACAC TACCATAAGTGAAACCCGGAAACGCAAAGGAAAGCAAGTATATCTTGGTGGATATGAAACTGAAGAGAAAGCAGCACAAG CATATGACCTTGCAGCTTTAAAGTTGTGGGGAGCAGGAGCACATTCTAAAATGAACTTCCCAATTCCCAATTATGCAAAA GAAGTTGAGGAAATGAAAGACATGTCACGTGAGGAGTATATTAATTCTCTAAGAAGGAAAAGCATTTGCTTTGCAAGAGG CAATTCGATTTATAGAGGAGTCACAAGACAACATGAAGGTCGTCGATGGCAAGCTAGAATTGGTAGGGTGGCAGGTGGTA GAGATATCTATCTTGGAACATATGACACCGAGGAGGAAGCTGCAGAGGCGTATGATATTGCGGCCGTTAAACTTCGAGGA ATGAGTGCCATTACCAATTTTCATATCAGTAACTACTACAAGGAAGGCTCAAAGGAACTGGATATTGTCCCTATAGACGT TACAGAGAAAGAGCTTCCGAAAGTGAAAATTAGAAGATTCATAATGCCGTGA |
| Microexon DNA seq | TATATCTTG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAACACTACCATAAGTGAAACCCGGAAACGCAAAGGAAAGCAAGTATATCTTGGTGGATATGAAACTGAAGAGAAAGCAGCACAAGCATATGACCTTGCAGCT |
| Microexon-tag Amino Acid seq | WDNTTISETRKRKGKQVYLGGYETEEKAAQAYDLAA |
| Transcript ID | XM_026566716.1 |
| Gene ID | Ps.59806 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 4.9e-11 |
| Motif start | 82 |
| Motif end | 144 |
| Protein seq | >XM_026566716.1 MGTLIPASENEICLLNQEVSLIDQVSIVADYERLPSSNDSSSKLIPPPSSSSSVNVNPASSSATGNEQVKKALYKVGQRT SNFRGVTSLFCRHRWTGKFEAHLWDNTTISETRKRKGKQVYLGGYETEEKAAQAYDLAALKLWGAGAHSKMNFPIPNYAK EVEEMKDMSREEYINSLRRKSICFARGNSIYRGVTRQHEGRRWQARIGRVAGGRDIYLGTYDTEEEAAEAYDIAAVKLRG MSAITNFHISNYYKEGSKELDIVPIDVTEKELPKVKIRRFIMP* |
| CDS seq | >XM_026566716.1 ATGGGAACTCTGATTCCTGCTTCAGAAAATGAGATATGTTTATTAAACCAGGAGGTATCCCTTATAGACCAAGTTTCAAT AGTGGCAGACTATGAGAGATTACCTTCTTCCAATGATTCATCTTCTAAATTAATTCCACCACCATCAAGTAGCAGTAGTG TTAATGTAAATCCTGCAAGCAGTAGTGCAACTGGAAACGAACAAGTAAAGAAGGCATTATATAAAGTTGGTCAGAGAACA TCGAATTTTCGTGGAGTTACTAGTTTGTTCTGCAGACACAGATGGACTGGCAAATTTGAAGCACATCTTTGGGATAACAC TACCATAAGTGAAACCCGGAAACGCAAAGGAAAGCAAGTATATCTTGGTGGATATGAAACTGAAGAGAAAGCAGCACAAG CATATGACCTTGCAGCTTTAAAGTTGTGGGGAGCAGGAGCACATTCTAAAATGAACTTCCCAATTCCCAATTATGCAAAA GAAGTTGAGGAAATGAAAGACATGTCACGTGAGGAGTATATTAATTCTCTAAGAAGGAAAAGCATTTGCTTTGCAAGAGG CAATTCGATTTATAGAGGAGTCACAAGACAACATGAAGGTCGTCGATGGCAAGCTAGAATTGGTAGGGTGGCAGGTGGTA GAGATATCTATCTTGGAACATATGACACCGAGGAGGAAGCTGCAGAGGCGTATGATATTGCGGCCGTTAAACTTCGAGGA ATGAGTGCCATTACCAATTTTCATATCAGTAACTACTACAAGGAAGGCTCAAAGGAACTGGATATTGTCCCTATAGACGT TACAGAGAAAGAGCTTCCGAAAGTGAAAATTAGAAGATTCATAATGCCGTGA |