
| Microexon ID | Ps_NC_039359.1:53674527-53674535:- |
| Species | Papaver somniferum | Coordinates | NC_039359.1:53674527..53674535 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 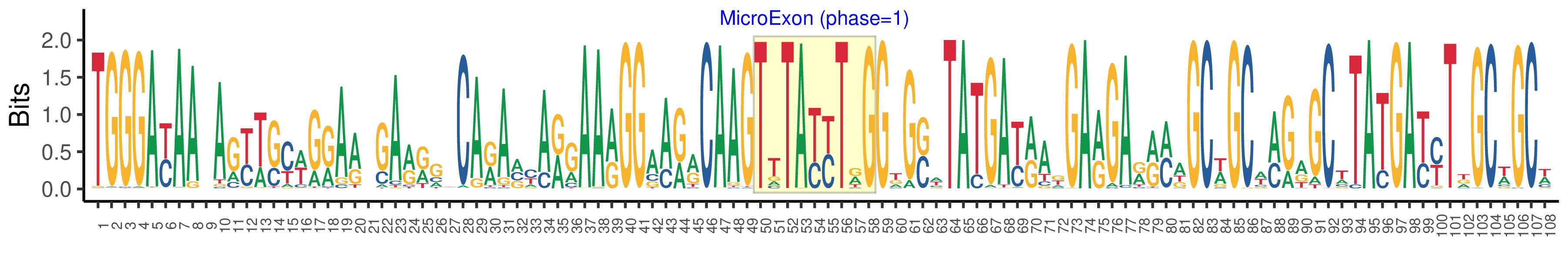 |
| Alignment of exons | 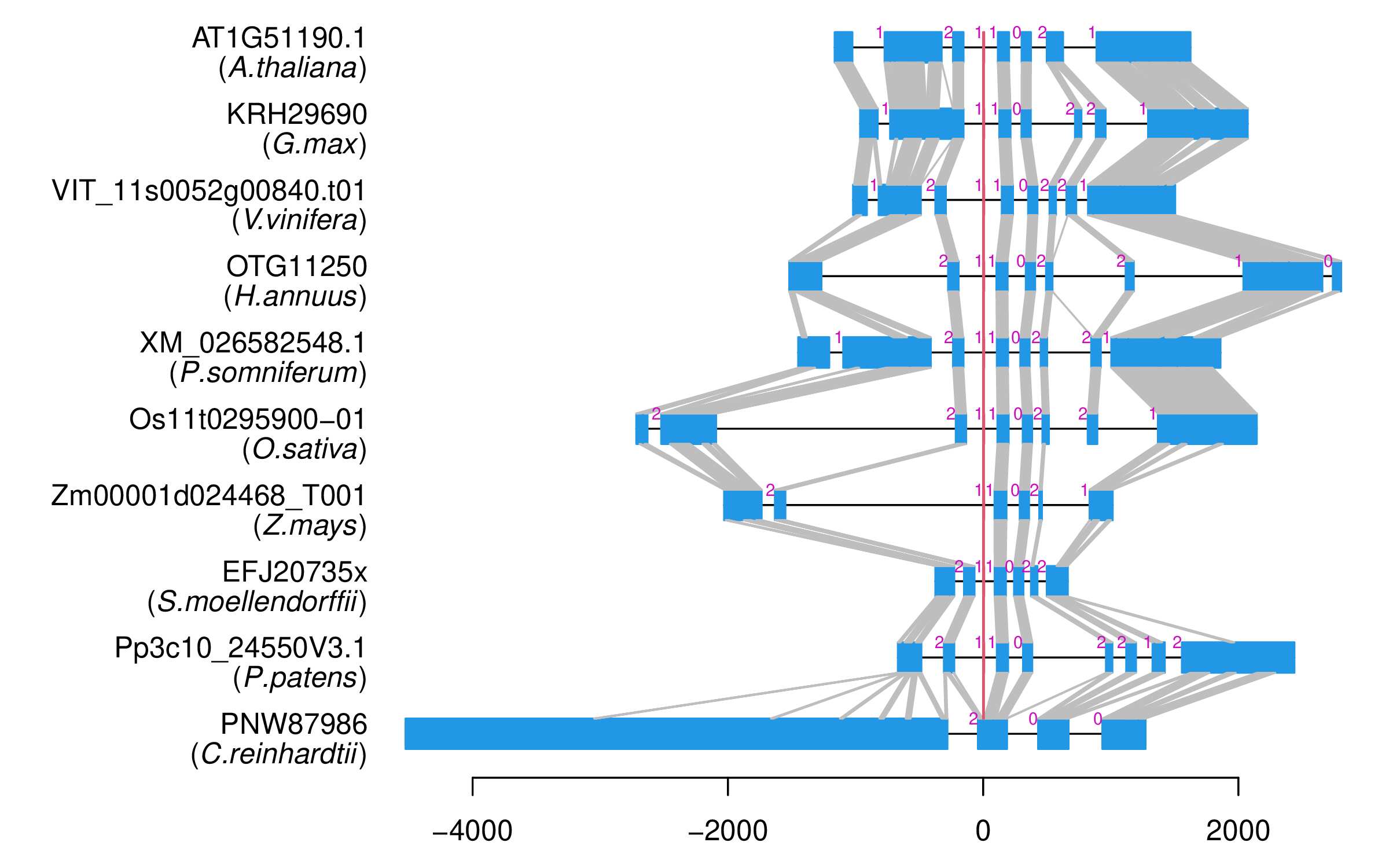 |
Ps_NC_039359.1:53674527-53674535:- does not have available information here.
| Transcript ID | XM_026596951.1 |
| Protein ID | XP_026452736.1 |
| Gene ID | LOC113353301 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 9.1e-10 |
| Motif start | 111 |
| Motif end | 168 |
| Protein seq | >XP_026452736.1 MEAGDSKSRRKISPRLLDLNSEWAVEEERRLFGGIEERKLELKLGPPNEEEKGFSNGSNRKNIVKSPSSNTLNSTVNKTP ISLLFTSTLTSTTTTNNTFHYNYKKKQKISQYRGVTRDVRKGIHKAQIWDGSCRKEGIRKGKQVYLGKYDEEDAAAKAYD LAALKYFGPTARINFPLANYETEIKEMEC* |
| CDS seq | >XM_026596951.1 ATGGAGGCTGGAGATTCGAAAAGTCGCCGAAAAATTAGCCCACGGCTGCTGGATTTGAATTCAGAATGGGCGGTAGAAGA AGAAAGAAGATTGTTTGGTGGTATAGAGGAAAGAAAACTGGAACTAAAACTCGGACCACCTAATGAAGAAGAGAAAGGGT TTTCTAATGGAAGCAATAGAAAGAATATAGTTAAATCTCCTTCTTCTAATACCCTTAATAGTACTGTAAACAAAACACCT ATTTCGCTTCTATTTACTTCAACTTTGACTAGCACTACTACTACGAATAATACTTTCCACTACAACTATAAAAAGAAGCA AAAAATTTCTCAATACCGTGGTGTTACTAGAGATGTGCGGAAAGGAATACATAAAGCGCAAATCTGGGATGGAAGTTGTA GAAAAGAAGGTATAAGAAAAGGAAAACAAGTTTATCTTGGTAAATATGATGAGGAAGATGCAGCAGCCAAGGCTTATGAT TTAGCTGCATTGAAGTATTTTGGCCCTACTGCTCGTATAAATTTCCCTTTAGCTAATTATGAGACAGAAATTAAAGAAAT GGAGTGTTGA |
| Microexon DNA seq | TTTATCTTG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | ATCTGGGATGGAAGTTGTAGAAAAGAAGGTATAAGAAAAGGAAAACAAGTTTATCTTGGTAAATATGATGAGGAAGATGCAGCAGCCAAGGCTTATGATTTAGCTGCA |
| Microexon-tag Amino Acid seq | IWDGSCRKEGIRKGKQVYLGKYDEEDAAAKAYDLAA |
| Transcript ID | Ps.10011.1 |
| Gene ID | Ps.10011 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 9.1e-10 |
| Motif start | 111 |
| Motif end | 168 |
| Protein seq | >Ps.10011.1 MEAGDSKSRRKISPRLLDLNSEWAVEEERRLFGGIEERKLELKLGPPNEEEKGFSNGSNRKNIVKSPSSNTLNSTVNKTP ISLLFTSTLTSTTTTNNTFHYNYKKKQKISQYRGVTRDVRKGIHKAQIWDGSCRKEGIRKGKQVYLGKYDEEDAAAKAYD LAALKYFGPTARINFPLANYETEIKEMEC* |
| CDS seq | >Ps.10011.1 ATGGAGGCTGGAGATTCGAAAAGTCGCCGAAAAATTAGCCCACGGCTGCTGGATTTGAATTCAGAATGGGCGGTAGAAGA AGAAAGAAGATTGTTTGGTGGTATAGAGGAAAGAAAACTGGAACTAAAACTCGGACCACCTAATGAAGAAGAGAAAGGGT TTTCTAATGGAAGCAATAGAAAGAATATAGTTAAATCTCCTTCTTCTAATACCCTTAATAGTACTGTAAACAAAACACCT ATTTCGCTTCTATTTACTTCAACTTTGACTAGCACTACTACTACGAATAATACTTTCCACTACAACTATAAAAAGAAGCA AAAAATTTCTCAATACCGTGGTGTTACTAGAGATGTGCGGAAAGGAATACATAAAGCGCAAATCTGGGATGGAAGTTGTA GAAAAGAAGGTATAAGAAAAGGAAAACAAGTTTATCTTGGTAAATATGATGAGGAAGATGCAGCAGCCAAGGCTTATGAT TTAGCTGCATTGAAGTATTTTGGCCCTACTGCTCGTATAAATTTCCCTTTAGCTAATTATGAGACAGAAATTAAAGAAAT GGAGTGTTGA |