
| Microexon ID | Gm_KZ847154:6999-7007:+ |
| Species | Glycine max | Coordinates | KZ847154:6999..7007 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 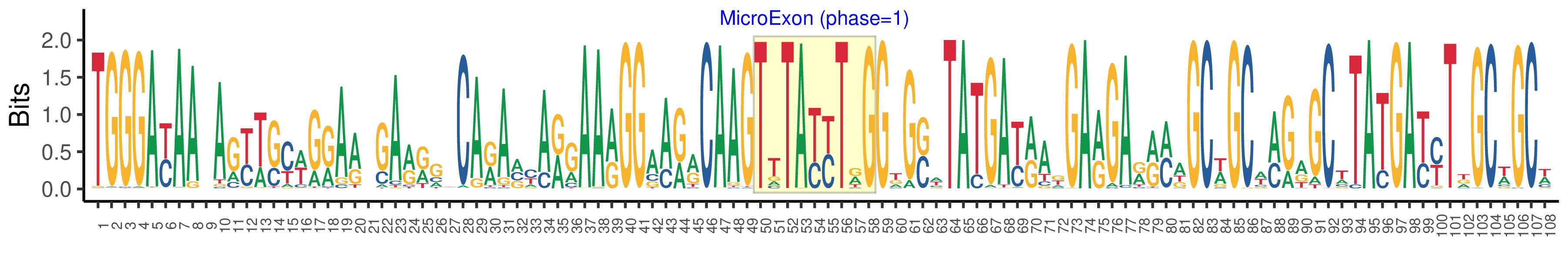 |
| Alignment of exons |  |
| Microexon DNA seq | TGTATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAACAGTTGCAAGAAGGAAGGGCAAACAAGGAAAGGACGACAAGTGTATTTGGGTGGTTATGATATGGAAGAGAAAGCTGCAAGGGCTTATGATCTTGCGGCT |
| Microexon-tag Amino Acid Seq | WDNSCKKEGQTRKGRQVYLGGYDMEEKAARAYDLAA |
| Microexon-tag spanning region | 6806-7163 |
| Microexon-tag prediction score | 0.9796 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | RCH53562x |
| Reference Transcript ID | RCH53562 |
| Gene ID | GLYMA_U037700 |
| Gene Name | NA |
| Transcript ID | RCH53562 |
| Protein ID | RCH53562 |
| Gene ID | GLYMA_U037700 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 1.1e-13 |
| Motif start | 352 |
| Motif end | 410 |
| Protein seq | >RCH53562 MKSMNDSNTVDDGNNHNNWLGFSLSPHMKMDVVTSSTTTGPHHPHQHHHHHHYYHHPHEASAAACNNNNNTVPTNFYMSP SHLNTSGICYGVGENSAFHTPLAMMPLKSDGSLCIMEALTRSQTQMMVPTSSPKLEDFLGGATMGAQDYGTHEREAMALS LDSIYYSNQNAEPETNRDHSSSLDLLSDHFRHQTHHHPYYSGLGIYQVEEEETKEQPHVAVCSSQMPQVVEGSIACFKNW VPTREYSSSSTQQNLEQHQVNSSSSGGLGEDNNVAYGNVGVGSSVGCGELQSLSLSMSPGSQSSCVTVPTQISSSGTDSV AVDAKKRGSSKLGQKQPVHRKSIDTFGQRTSQYRGVTRHRWTGRYEAHLWDNSCKKEGQTRKGRQVYLGGYDMEEKAARA YDLAALKYWGPSTHINFPLENYQTQLEEMKNMSRQEYVAHLRRKSSGFSRGASMYRGVTRHHQHGRWQARIGRVAGNKDL YLGTFSTQEEAAEAYDVAAIKFRGVNAVTNFDISRYDVERIMASNTLLAES* |
| CDS seq | >RCH53562 ATGAAGTCCATGAATGATAGTAACACCGTTGATGATGGGAACAATCATAATAACTGGTTGGGATTCTCTCTCTCACCCCA CATGAAAATGGATGTTGTTACTTCTTCTACTACCACTGGTCCTCATCATCCCCACCAACACCATCATCATCATCACTACT ATCATCACCCTCACGAGGCTTCTGCTGCAGCTTGCAACAACAACAACAACACTGTTCCCACTAACTTCTATATGTCACCC TCGCACCTCAACACCTCTGGAATATGTTATGGTGTTGGAGAAAACAGTGCCTTTCACACTCCTTTGGCCATGATGCCTCT CAAGTCAGATGGGTCACTTTGCATTATGGAGGCTCTAACAAGATCACAAACCCAAATGATGGTGCCAACTTCATCTCCAA AACTTGAGGACTTTCTAGGTGGTGCAACTATGGGGGCTCAAGACTATGGAACCCATGAGAGAGAAGCAATGGCTCTAAGC CTAGACAGTATCTACTACAGCAACCAGAATGCTGAACCTGAAACCAACAGGGACCATTCATCTTCTCTTGACCTTCTTTC TGACCATTTCAGGCACCAAACCCATCATCACCCATATTACTCAGGACTTGGGATTTACCAAGTGGAGGAAGAAGAAACCA AGGAACAACCACACGTTGCAGTTTGCAGCTCCCAAATGCCTCAAGTGGTTGAAGGCAGCATTGCTTGCTTCAAAAACTGG GTGCCAACAAGGGAATACTCTTCTTCTTCCACTCAGCAGAATCTGGAGCAGCATCAAGTGAATAGTAGTAGCAGTGGTGG CCTTGGAGAGGATAATAATGTAGCTTATGGGAATGTTGGTGTTGGTAGTAGTGTTGGTTGTGGTGAGTTACAGTCTTTGA GTTTGTCTATGAGTCCTGGTTCTCAATCAAGCTGTGTCACTGTTCCAACTCAGATCTCATCTTCTGGAACTGACTCAGTT GCTGTGGATGCCAAAAAGAGAGGCTCTTCTAAGCTTGGACAGAAGCAACCTGTGCATAGGAAATCCATCGACACATTTGG TCAAAGAACTTCTCAGTATAGAGGTGTCACAAGGCATAGATGGACTGGTAGATATGAAGCACATTTGTGGGATAACAGTT GCAAGAAGGAAGGGCAAACAAGGAAAGGACGACAAGTGTATTTGGGTGGTTATGATATGGAAGAGAAAGCTGCAAGGGCT TATGATCTTGCGGCTCTCAAGTATTGGGGACCTTCAACACACATAAACTTCCCGCTAGAAAATTACCAAACTCAACTTGA AGAAATGAAGAATATGAGTAGGCAGGAATACGTGGCCCACTTGAGAAGAAAGAGTAGTGGGTTTTCAAGGGGTGCCTCAA TGTACAGAGGAGTGACAAGGCACCACCAACATGGCAGGTGGCAAGCAAGGATAGGCAGAGTTGCAGGAAATAAGGACCTT TATCTTGGGACATTCAGCACTCAAGAGGAAGCAGCTGAAGCATATGATGTAGCTGCAATCAAATTTCGTGGGGTGAATGC TGTCACCAACTTTGACATATCCAGATACGACGTTGAGAGAATAATGGCCAGCAACACCCTTCTAGCTGAGAGCTAG |
| Microexon DNA seq | TGTATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAACAGTTGCAAGAAGGAAGGGCAAACAAGGAAAGGACGACAAGTGTATTTGGGTGGTTATGATATGGAAGAGAAAGCTGCAAGGGCTTATGATCTTGCGGCT |
| Microexon-tag Amino Acid seq | WDNSCKKEGQTRKGRQVYLGGYDMEEKAARAYDLAA |
| Transcript ID | RCH53564 |
| Gene ID | Gm.54782 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 1.5e-13 |
| Motif start | 352 |
| Motif end | 410 |
| Protein seq | >RCH53564 MKSMNDSNTVDDGNNHNNWLGFSLSPHMKMDVVTSSTTTGPHHPHQHHHHHHYYHHPHEASAAACNNNNNTVPTNFYMSP SHLNTSGICYGVGENSAFHTPLAMMPLKSDGSLCIMEALTRSQTQMMVPTSSPKLEDFLGGATMGAQDYGTHEREAMALS LDSIYYSNQNAEPETNRDHSSSLDLLSDHFRHQTHHHPYYSGLGIYQVEEEETKEQPHVAVCSSQMPQVVEGSIACFKNW VPTREYSSSSTQQNLEQHQVNSSSSGGLGEDNNVAYGNVGVGSSVGCGELQSLSLSMSPGSQSSCVTVPTQISSSGTDSV AVDAKKRGSSKLGQKQPVHRKSIDTFGQRTSQYRGVTRHRWTGRYEAHLWDNSCKKEGQTRKGRQVYLGGYDMEEKAARA YDLAALKYWGPSTHINFPLENYQTQLEEMKNMSRQEYVAHLRRKSSGFSRGASMYRGVTRHHQHGRWQARIGRVAGNKDL YLGTFSTQEEAAEAYDVAAIKFRGVNAVTNFDISRYDMSLYHHQQQSNNCDQKTIKCENYNRGGAAFSVSLQDLIGIDSV GSSQGMMDESTKIGTHFSNPSSLVTSLSSSREGSPDKMGPTLLIPKPPMGSKIVTSPTVANGVTVGSWFPSQMRPVSMSH LPVFAAWSDA* |
| CDS seq | >RCH53564 ATGAAGTCCATGAATGATAGTAACACCGTTGATGATGGGAACAATCATAATAACTGGTTGGGATTCTCTCTCTCACCCCA CATGAAAATGGATGTTGTTACTTCTTCTACTACCACTGGTCCTCATCATCCCCACCAACACCATCATCATCATCACTACT ATCATCACCCTCACGAGGCTTCTGCTGCAGCTTGCAACAACAACAACAACACTGTTCCCACTAACTTCTATATGTCACCC TCGCACCTCAACACCTCTGGAATATGTTATGGTGTTGGAGAAAACAGTGCCTTTCACACTCCTTTGGCCATGATGCCTCT CAAGTCAGATGGGTCACTTTGCATTATGGAGGCTCTAACAAGATCACAAACCCAAATGATGGTGCCAACTTCATCTCCAA AACTTGAGGACTTTCTAGGTGGTGCAACTATGGGGGCTCAAGACTATGGAACCCATGAGAGAGAAGCAATGGCTCTAAGC CTAGACAGTATCTACTACAGCAACCAGAATGCTGAACCTGAAACCAACAGGGACCATTCATCTTCTCTTGACCTTCTTTC TGACCATTTCAGGCACCAAACCCATCATCACCCATATTACTCAGGACTTGGGATTTACCAAGTGGAGGAAGAAGAAACCA AGGAACAACCACACGTTGCAGTTTGCAGCTCCCAAATGCCTCAAGTGGTTGAAGGCAGCATTGCTTGCTTCAAAAACTGG GTGCCAACAAGGGAATACTCTTCTTCTTCCACTCAGCAGAATCTGGAGCAGCATCAAGTGAATAGTAGTAGCAGTGGTGG CCTTGGAGAGGATAATAATGTAGCTTATGGGAATGTTGGTGTTGGTAGTAGTGTTGGTTGTGGTGAGTTACAGTCTTTGA GTTTGTCTATGAGTCCTGGTTCTCAATCAAGCTGTGTCACTGTTCCAACTCAGATCTCATCTTCTGGAACTGACTCAGTT GCTGTGGATGCCAAAAAGAGAGGCTCTTCTAAGCTTGGACAGAAGCAACCTGTGCATAGGAAATCCATCGACACATTTGG TCAAAGAACTTCTCAGTATAGAGGTGTCACAAGGCATAGATGGACTGGTAGATATGAAGCACATTTGTGGGATAACAGTT GCAAGAAGGAAGGGCAAACAAGGAAAGGACGACAAGTGTATTTGGGTGGTTATGATATGGAAGAGAAAGCTGCAAGGGCT TATGATCTTGCGGCTCTCAAGTATTGGGGACCTTCAACACACATAAACTTCCCGCTAGAAAATTACCAAACTCAACTTGA AGAAATGAAGAATATGAGTAGGCAGGAATACGTGGCCCACTTGAGAAGAAAGAGTAGTGGGTTTTCAAGGGGTGCCTCAA TGTACAGAGGAGTGACAAGGCACCACCAACATGGCAGGTGGCAAGCAAGGATAGGCAGAGTTGCAGGAAATAAGGACCTT TATCTTGGGACATTCAGCACTCAAGAGGAAGCAGCTGAAGCATATGATGTAGCTGCAATCAAATTTCGTGGGGTGAATGC TGTCACCAACTTTGACATATCCAGATACGACATGAGTTTGTATCATCATCAGCAACAGTCAAACAACTGTGACCAGAAAA CCATCAAGTGTGAAAATTATAATAGAGGTGGTGCTGCTTTCTCTGTGTCCCTACAAGATCTCATTGGGATTGACTCAGTA GGATCTAGCCAAGGCATGATGGATGAGTCTACTAAGATAGGGACTCATTTTTCAAACCCTTCCTCGCTGGTCACCAGTTT AAGCAGCTCAAGGGAAGGTAGCCCTGATAAAATGGGCCCCACTTTGCTCATTCCAAAGCCTCCAATGGGGTCAAAGATTG TTACTAGCCCTACTGTTGCCAATGGTGTCACTGTTGGCTCTTGGTTTCCCTCTCAAATGAGGCCAGTCTCAATGTCTCAC TTGCCAGTTTTTGCTGCTTGGAGTGATGCCTAG |