
| Microexon ID | Gm_9:46285136-46285144:- |
| Species | Glycine max | Coordinates | 9:46285136..46285144 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 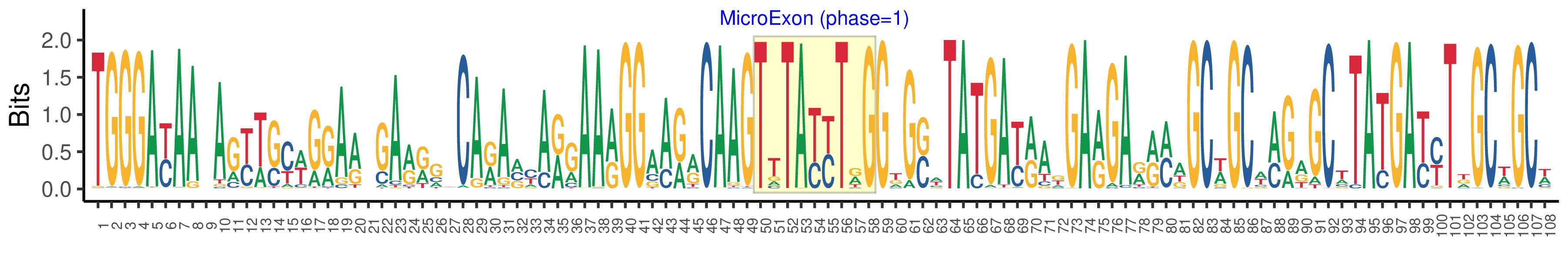 |
| Alignment of exons |  |
| Microexon DNA seq | TATACCTTG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAAGCTTTCTTGGAACATAACACAAAAGAAGAAAGGGAAGCAAGTATACCTTGGGGCTTATGATGAAGAAGAATCTGCTGCCAGAGCATATGATTTAGCTGCA |
| Microexon-tag Amino Acid Seq | WDKLSWNITQKKKGKQVYLGAYDEEESAARAYDLAA |
| Microexon-tag spanning region | 46284990-46285339 |
| Microexon-tag prediction score | 0.9308 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH40131x |
| Reference Transcript ID | KRH40131 |
| Gene ID | GLYMA_09G240400 |
| Gene Name | NA |
| Transcript ID | KRH40131 |
| Protein ID | KRH40131 |
| Gene ID | GLYMA_09G240400 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 1.7e-13 |
| Motif start | 71 |
| Motif end | 130 |
| Protein seq | >KRH40131 MAMMKENIIEVSLGRRQMSMTEGEFQGTRSVKRRRREVAAAAGSGDDNHQQQLPQQEVGENTTVNTTKRSSRFRGVSRHR WTGRYEAHLWDKLSWNITQKKKGKQVYLGAYDEEESAARAYDLAALKYWGTSTFTNFPISDYEKEIQIMQTMTKEEYLAT LRRKSSGFSRGVSKYRGVARHHHNGRWEARIGRVFGNKYLYLGTYSTQEEAARAYDIAAIEYRGIHAVTNFDLSTYIKWL KPSGGGTLEANLESHAALEHQKVASPSNYALTEESKSLALHNSFFSPYSLDSPVKHERFGNKTYQFSSNKSSSPTALGLL LRSSLFRELVEKNSNVSGEEDDGEATKDQQTQIATDDDLGGIFFDSFSDIPFVCDPNRYDLELQERDLHSIF* |
| CDS seq | >KRH40131 ATGGCAATGATGAAAGAGAATATTATTGAAGTGAGCCTTGGAAGGAGGCAAATGTCTATGACAGAAGGTGAGTTCCAAGG AACACGGAGCGTCAAGAGACGACGAAGAGAGGTTGCTGCAGCCGCAGGCAGTGGTGATGATAACCACCAACAGCAGTTAC CACAACAAGAAGTTGGTGAAAATACTACAGTCAACACTACAAAGAGAAGCTCAAGATTTCGCGGTGTTAGCAGACATAGA TGGACGGGTCGATATGAAGCTCACTTGTGGGACAAGCTTTCTTGGAACATAACACAAAAGAAGAAAGGGAAGCAAGTATA CCTTGGGGCTTATGATGAAGAAGAATCTGCTGCCAGAGCATATGATTTAGCTGCACTTAAGTATTGGGGAACTTCAACTT TCACAAATTTTCCAATATCAGATTATGAGAAAGAGATACAAATAATGCAGACTATGACCAAAGAGGAGTATTTGGCCACT TTAAGGAGAAAGAGCAGTGGCTTTTCAAGAGGTGTATCAAAGTATAGGGGTGTTGCGAGGCACCACCACAATGGTAGATG GGAAGCAAGAATAGGAAGGGTTTTTGGAAACAAATATCTCTACCTTGGAACCTACAGCACCCAAGAGGAAGCTGCGCGTG CTTATGACATAGCAGCCATTGAGTACAGAGGAATTCATGCTGTAACAAACTTTGATTTGAGCACTTACATAAAATGGTTA AAGCCTTCAGGAGGAGGCACCCTAGAAGCAAATCTTGAATCACATGCAGCACTAGAGCATCAAAAGGTGGCATCCCCATC TAATTATGCTCTAACAGAAGAGTCTAAGTCTTTGGCCCTTCACAACAGTTTTTTCAGTCCATATTCTCTGGATTCCCCTG TAAAGCATGAAAGATTTGGAAACAAAACCTACCAATTCTCAAGCAATAAGTCATCTTCTCCCACTGCACTTGGTCTCCTT CTCCGCTCTTCGTTATTTAGGGAATTAGTTGAAAAGAATTCAAATGTTTCTGGAGAAGAAGATGATGGGGAAGCCACAAA GGATCAACAGACACAAATAGCTACTGATGATGATCTGGGTGGAATCTTCTTCGATAGCTTTAGTGATATTCCATTTGTGT GTGACCCCAATAGATACGACTTGGAATTGCAAGAAAGAGACCTGCACTCAATATTTTGA |
Gm_9:46285136-46285144:- does not have available information here.