
| Microexon ID | Gm_8:37696730-37696738:- |
| Species | Glycine max | Coordinates | 8:37696730..37696738 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 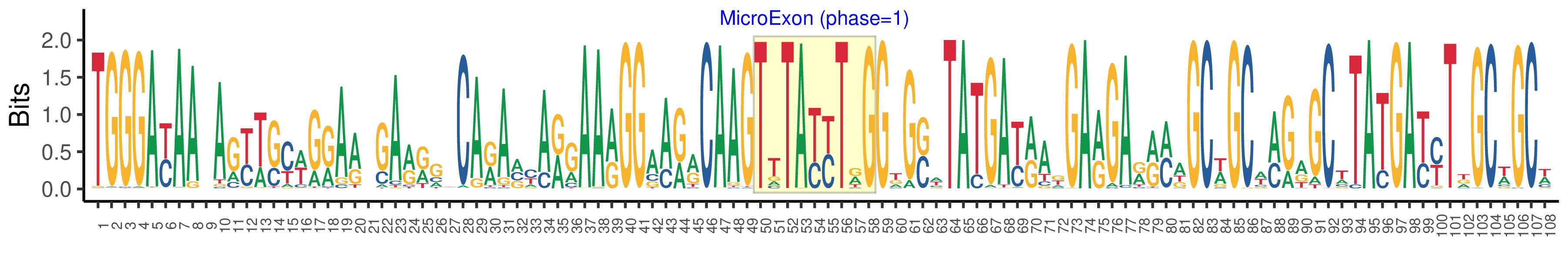 |
| Alignment of exons |  |
| Microexon DNA seq | TGTACTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAACAGCTGTAGGCGAGAGGGTCAGGCCAGGAAAGGGCGTCAAGTGTACTTGGGTGGATATGACAAGGAAGATAAGGCTGCGAGGGCTTATGATTTAGCAGCT |
| Microexon-tag Amino Acid Seq | WDNSCRREGQARKGRQVYLGGYDKEDKAARAYDLAA |
| Microexon-tag spanning region | 37696575-37698972 |
| Microexon-tag prediction score | 0.9678 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH45548x |
| Reference Transcript ID | KRH45548 |
| Gene ID | GLYMA_08G279000 |
| Gene Name | NA |
| Transcript ID | KRH45548 |
| Protein ID | KRH45548 |
| Gene ID | GLYMA_08G279000 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 5e-12 |
| Motif start | 164 |
| Motif end | 222 |
| Protein seq | >KRH45548 MAGATNWLTFSLTHMEMQQFGPYDTTTHSHCFNDSNFYGWTNSKSVMDSQSETQQLAVPKMEDFYFGDHQQDLKAMIVPG FKALSGTNSGSEVDDSTSNKTVTRVAPAALGGAHSGESNCKGSALTLCDVAANGSDDSNNKAVVAVGFDTRKKVAHTFGQ RTSIYRGVTRHRWTGRYEAHLWDNSCRREGQARKGRQVYLGGYDKEDKAARAYDLAALKYWGPTATTNFPELEEMEHAGR QEFIASLRRKSSGFSRGASAYRGVTRHHQQGRWQARIGRVAGNKDLYLGTFSTEEEAAEAYDIAAIKFRGSSAVTNFEMS RYDVDTILNNSLPVGGVAKRLKVSPATENKALISTIIQPPQNTNASSSINFSAIQPVSSIPYDSAITCYPHNFFHHHFNP TNGGTSSSDSAAAAAVTDAIALNTLPPPAVPGFFIWPHQSY* |
| CDS seq | >KRH45548 ATGGCTGGTGCCACGAATTGGCTTACGTTTTCGTTGACTCACATGGAAATGCAGCAGTTCGGTCCATACGACACCACCAC TCACTCTCACTGCTTCAACGACAGCAATTTTTATGGGTGGACGAATTCTAAGAGCGTCATGGATTCACAGAGTGAGACCC AACAACTTGCGGTGCCAAAAATGGAAGATTTTTACTTCGGTGATCACCAGCAAGATCTGAAGGCGATGATTGTTCCTGGG TTCAAAGCGTTGTCTGGGACCAACTCGGGGTCCGAGGTGGATGACTCGACCTCCAACAAGACGGTGACTCGGGTTGCACC CGCTGCGTTGGGCGGTGCTCACTCTGGCGAATCTAATTGTAAAGGATCAGCTTTGACTTTATGCGATGTTGCTGCGAATG GTTCTGATGATAGTAATAATAAGGCCGTTGTTGCCGTGGGGTTTGATACTCGGAAGAAGGTTGCTCATACGTTTGGCCAG CGAACTTCAATTTACAGAGGGGTCACAAGGCACCGATGGACGGGAAGATATGAAGCGCATCTATGGGATAACAGCTGTAG GCGAGAGGGTCAGGCCAGGAAAGGGCGTCAAGTGTACTTGGGTGGATATGACAAGGAAGATAAGGCTGCGAGGGCTTATG ATTTAGCAGCTCTAAAGTACTGGGGACCAACAGCCACGACTAACTTTCCTGAATTGGAGGAGATGGAACATGCAGGAAGG CAAGAATTTATTGCATCACTGCGAAGGAAAAGTAGTGGTTTTTCAAGGGGAGCTTCTGCTTACAGAGGTGTTACAAGGCA TCACCAACAGGGTAGATGGCAAGCAAGAATTGGCCGAGTTGCGGGAAACAAGGATCTATACTTGGGAACATTTTCAACTG AAGAGGAGGCAGCAGAGGCTTATGACATTGCAGCCATAAAGTTCAGAGGTTCGAGCGCCGTAACCAATTTCGAGATGAGT CGATATGACGTCGACACTATATTGAACAACTCGCTTCCAGTTGGTGGCGTAGCAAAGCGCTTGAAAGTTTCCCCAGCAAC GGAGAACAAAGCTCTTATCAGCACCATTATTCAACCACCTCAGAACACAAACGCAAGTAGCAGCATCAATTTCTCAGCCA TTCAGCCAGTTTCTTCTATACCCTATGATTCTGCAATCACATGTTACCCCCACAACTTCTTCCACCACCATTTCAATCCT ACCAACGGTGGAACTAGCTCATCAGATTCTGCTGCTGCTGCAGCAGTGACCGATGCGATTGCACTCAATACCCTTCCACC ACCAGCAGTTCCTGGGTTCTTCATATGGCCTCACCAGTCCTACTGA |
| Microexon DNA seq | TGTACTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAACAGCTGTAGGCGAGAGGGTCAGGCCAGGAAAGGGCGTCAAGTGTACTTGGGTGGATATGACAAGGAAGATAAGGCTGCGAGGGCTTATGATTTAGCAGCT |
| Microexon-tag Amino Acid seq | WDNSCRREGQARKGRQVYLGGYDKEDKAARAYDLAA |
| Transcript ID | Gm.50886.1 |
| Gene ID | Gm.50886 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 4.4e-12 |
| Motif start | 118 |
| Motif end | 176 |
| Protein seq | >Gm.50886.1 MDSQSETQQLAVPKMEDFYFGDHQQDLKAMIVPGFKALSGTNSGSEVDDSTSNKTVTRVAPAALGGAHSGESNCKGSALT LCDVAANGSDDSNNKAVVAVGFDTRKKVAHTFGQRTSIYRGVTRHRWTGRYEAHLWDNSCRREGQARKGRQVYLGGYDKE DKAARAYDLAALKYWGPTATTNFPISNYTKELEEMEHAGRQEFIASLRRKSSGFSRGASAYRGVTRHHQQGRWQARIGRV AGNKDLYLGTFSTEEEAAEAYDIAAIKFRGSSAVTNFEMSRYDVDTILNNSLPVGGVAKRLKVSPATENKALISTIIQPP QNTNASSSINFSAIQPVSSIPYDSAITCYPHNFFHHHFNPTNGGTSSSDSAAAAAVTDAIALNTLPPPAVPGFFIWPHQS Y* |
| CDS seq | >Gm.50886.1 ATGGATTCACAGAGTGAGACCCAACAACTTGCGGTGCCAAAAATGGAAGATTTTTACTTCGGTGATCACCAGCAAGATCT GAAGGCGATGATTGTTCCTGGGTTCAAAGCGTTGTCTGGGACCAACTCGGGGTCCGAGGTGGATGACTCGACCTCCAACA AGACGGTGACTCGGGTTGCACCCGCTGCGTTGGGCGGTGCTCACTCTGGCGAATCTAATTGTAAAGGATCAGCTTTGACT TTATGCGATGTTGCTGCGAATGGTTCTGATGATAGTAATAATAAGGCCGTTGTTGCCGTGGGGTTTGATACTCGGAAGAA GGTTGCTCATACGTTTGGCCAGCGAACTTCAATTTACAGAGGGGTCACAAGGCACCGATGGACGGGAAGATATGAAGCGC ATCTATGGGATAACAGCTGTAGGCGAGAGGGTCAGGCCAGGAAAGGGCGTCAAGTGTACTTGGGTGGATATGACAAGGAA GATAAGGCTGCGAGGGCTTATGATTTAGCAGCTCTAAAGTACTGGGGACCAACAGCCACGACTAACTTTCCTATTTCCAA TTATACAAAGGAATTGGAGGAGATGGAACATGCAGGAAGGCAAGAATTTATTGCATCACTGCGAAGGAAAAGTAGTGGTT TTTCAAGGGGAGCTTCTGCTTACAGAGGTGTTACAAGGCATCACCAACAGGGTAGATGGCAAGCAAGAATTGGCCGAGTT GCGGGAAACAAGGATCTATACTTGGGAACATTTTCAACTGAAGAGGAGGCAGCAGAGGCTTATGACATTGCAGCCATAAA GTTCAGAGGTTCGAGCGCCGTAACCAATTTCGAGATGAGTCGATATGACGTCGACACTATATTGAACAACTCGCTTCCAG TTGGTGGCGTAGCAAAGCGCTTGAAAGTTTCCCCAGCAACGGAGAACAAAGCTCTTATCAGCACCATTATTCAACCACCT CAGAACACAAACGCAAGTAGCAGCATCAATTTCTCAGCCATTCAGCCAGTTTCTTCTATACCCTATGATTCTGCAATCAC ATGTTACCCCCACAACTTCTTCCACCACCATTTCAATCCTACCAACGGTGGAACTAGCTCATCAGATTCTGCTGCTGCTG CAGCAGTGACCGATGCGATTGCACTCAATACCCTTCCACCACCAGCAGTTCCTGGGTTCTTCATATGGCCTCACCAGTCC TACTGA |