
| Microexon ID | Gm_8:18544205-18544213:- |
| Species | Glycine max | Coordinates | 8:18544205..18544213 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 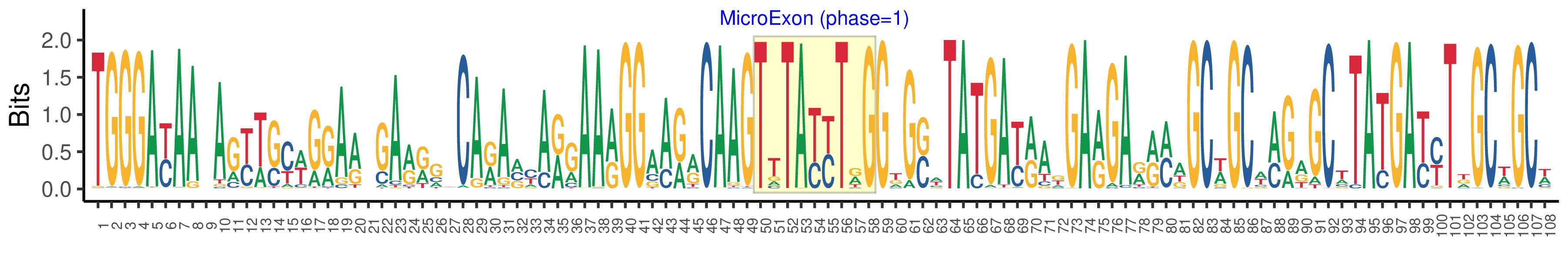 |
| Alignment of exons | 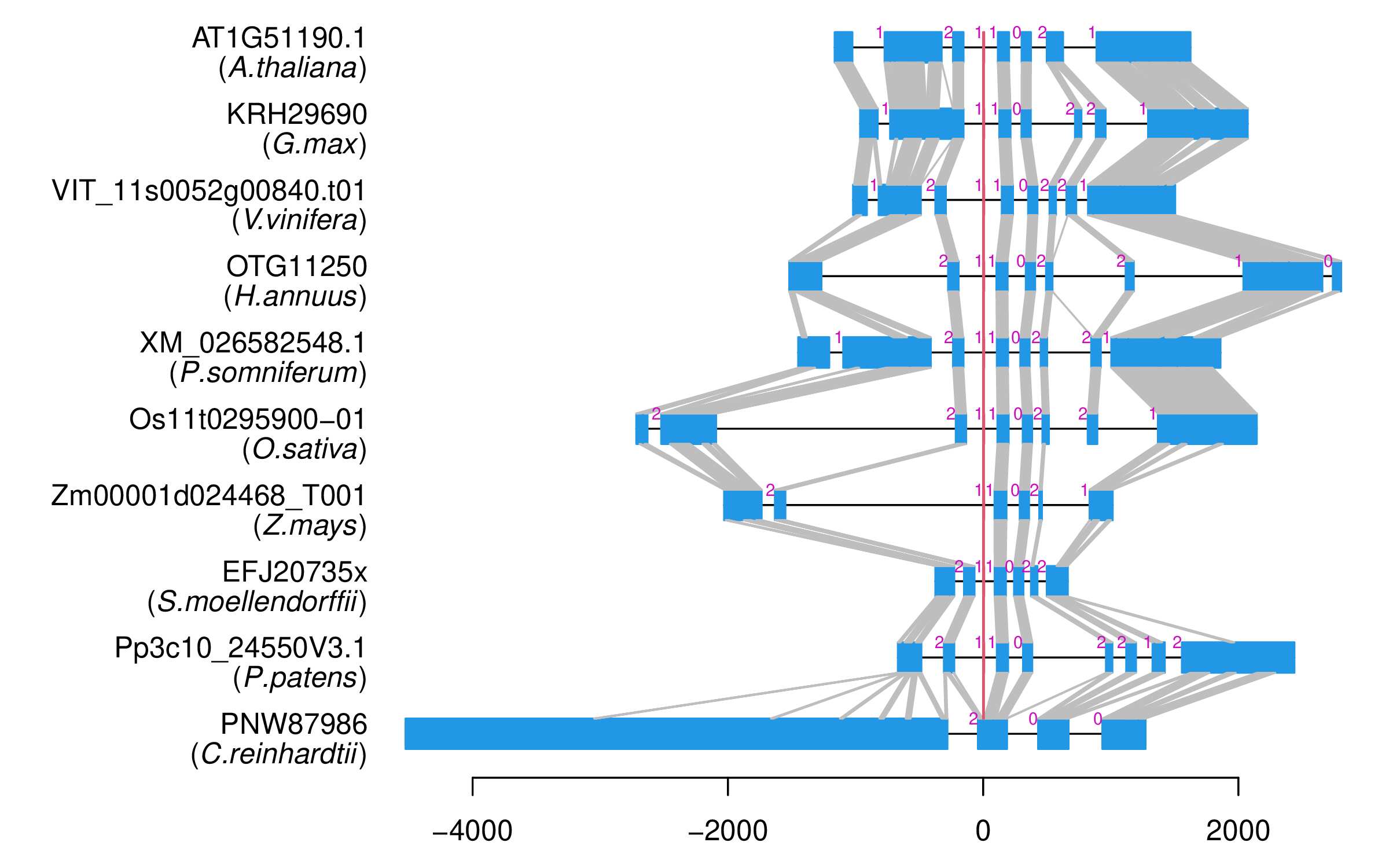 |
| Microexon DNA seq | TTTATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAAGAGCTCTTGGAACAACATTCAGAGCAAGAAGGGTAAACAAGTTTATTTGGGGGCATATGATACTGAAGAATCTGCAGCACGTACCTATGACCTTGCAGCC |
| Microexon-tag Amino Acid Seq | WDKSSWNNIQSKKGKQVYLGAYDTEESAARTYDLAA |
| Microexon-tag spanning region | 18543980-18544500 |
| Microexon-tag prediction score | 0.9447 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH44733x |
| Reference Transcript ID | KRH44733 |
| Gene ID | GLYMA_08G227700 |
| Gene Name | NA |
| Transcript ID | KRH44733 |
| Protein ID | KRH44733 |
| Gene ID | GLYMA_08G227700 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 2.8e-13 |
| Motif start | 56 |
| Motif end | 114 |
| Protein seq | >KRH44733 MKRSPASSCSSSTSSVGFEVHHPIEKRRPKHPRRNNLKSQKCKQNQTTTGGRRSSIYRGVTRHRWTGRFEAHLWDKSSWN NIQSKKGKQVYLGAYDTEESAARTYDLAALKYWGKDATLNFPIETYTKDLEEMDKVSREEYLASLRRQSSGFSRGISKYR GVARHHHNGRWEARIGRVCGNKYLYLGTYKTQEEAAVAYDMAAIEYRGVNAVTNFDISNYMDKIKKKNDQTLQQQQTEVQ TETVPNSSDSEEAEVEQQHTTTITTPPPSENLHMLPQEHQVQYTHHVTPRDEESSSLVTIMEHVLEQDLPWSFMYTGLSQ FQDPNLALSKGDDDLAGMFDGAGFEEDIDFLFSTQPGDHETESDVNNMSAVLDSVECGDTNGAGGRSMVYHHVDNNNKQK KMLSFASSSSPSSTTTTVSCDYALDL* |
| CDS seq | >KRH44733 ATGAAGAGGTCTCCAGCATCTTCTTGTTCATCATCCACTTCCTCTGTTGGGTTTGAAGTTCATCATCCCATTGAAAAAAG AAGGCCTAAGCATCCAAGGAGGAATAATTTGAAGTCACAAAAATGCAAGCAGAACCAAACCACCACTGGTGGCAGAAGAA GCTCTATCTATAGAGGAGTTACAAGGCATAGGTGGACAGGGAGGTTTGAAGCTCACCTATGGGATAAGAGCTCTTGGAAC AACATTCAGAGCAAGAAGGGTAAACAAGTTTATTTGGGGGCATATGATACTGAAGAATCTGCAGCACGTACCTATGACCT TGCAGCCCTTAAGTACTGGGGAAAAGATGCCACCCTGAATTTCCCGATAGAAACTTATACCAAGGACCTCGAGGAAATGG ACAAGGTTTCAAGAGAAGAATATTTAGCTTCTTTGCGGCGCCAAAGCAGTGGCTTTTCTAGAGGCATCTCTAAGTACCGT GGGGTTGCTAGGCATCATCATAATGGTCGCTGGGAAGCTCGCATTGGAAGAGTATGTGGAAACAAGTACCTCTACTTGGG AACATATAAAACTCAAGAGGAGGCAGCAGTGGCATATGACATGGCAGCAATTGAGTACCGTGGAGTCAATGCAGTGACCA ATTTTGACATAAGCAACTACATGGACAAAATAAAGAAGAAAAATGACCAAACCCTACAACAACAACAAACAGAAGTACAA ACAGAAACAGTTCCTAACTCCTCTGACTCTGAAGAAGCAGAAGTAGAACAACAACACACAACAACAATAACTACACCACC CCCATCTGAAAATCTGCACATGCTACCACAGGAACACCAAGTTCAATACACCCACCATGTCACTCCAAGGGATGAAGAAT CATCATCACTGGTCACAATTATGGAGCATGTGCTTGAACAGGATCTGCCATGGAGCTTCATGTACACTGGCTTGTCTCAG TTTCAAGATCCAAACTTGGCTTTAAGCAAAGGTGATGATGACTTGGCAGGCATGTTTGATGGTGCAGGGTTTGAGGAAGA CATTGATTTTCTGTTCAGCACACAACCTGGTGATCATGAGACTGAGAGTGATGTCAACAACATGAGTGCAGTTTTGGATA GTGTTGAGTGTGGAGACACAAATGGGGCTGGTGGAAGAAGCATGGTGTATCATCATGTGGATAATAATAATAAGCAGAAG AAGATGCTTTCATTTGCTTCTTCTTCTTCACCATCATCTACAACAACTACAGTTTCTTGTGACTATGCTCTAGATCTATG A |
| Microexon DNA seq | TTTATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAAGAGCTCTTGGAACAACATTCAGAGCAAGAAGGGTAAACAAGTTTATTTGGGGGCATATGATACTGAAGAATCTGCAGCACGTACCTATGACCTTGCAGCC |
| Microexon-tag Amino Acid seq | WDKSSWNNIQSKKGKQVYLGAYDTEESAARTYDLAA |
| Transcript ID | KRH44733 |
| Gene ID | Gm.50416 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 2.8e-13 |
| Motif start | 56 |
| Motif end | 114 |
| Protein seq | >KRH44733 MKRSPASSCSSSTSSVGFEVHHPIEKRRPKHPRRNNLKSQKCKQNQTTTGGRRSSIYRGVTRHRWTGRFEAHLWDKSSWN NIQSKKGKQVYLGAYDTEESAARTYDLAALKYWGKDATLNFPIETYTKDLEEMDKVSREEYLASLRRQSSGFSRGISKYR GVARHHHNGRWEARIGRVCGNKYLYLGTYKTQEEAAVAYDMAAIEYRGVNAVTNFDISNYMDKIKKKNDQTLQQQQTEVQ TETVPNSSDSEEAEVEQQHTTTITTPPPSENLHMLPQEHQVQYTHHVTPRDEESSSLVTIMEHVLEQDLPWSFMYTGLSQ FQDPNLALSKGDDDLAGMFDGAGFEEDIDFLFSTQPGDHETESDVNNMSAVLDSVECGDTNGAGGRSMVYHHVDNNNKQK KMLSFASSSSPSSTTTTVSCDYALDL* |
| CDS seq | >KRH44733 ATGAAGAGGTCTCCAGCATCTTCTTGTTCATCATCCACTTCCTCTGTTGGGTTTGAAGTTCATCATCCCATTGAAAAAAG AAGGCCTAAGCATCCAAGGAGGAATAATTTGAAGTCACAAAAATGCAAGCAGAACCAAACCACCACTGGTGGCAGAAGAA GCTCTATCTATAGAGGAGTTACAAGGCATAGGTGGACAGGGAGGTTTGAAGCTCACCTATGGGATAAGAGCTCTTGGAAC AACATTCAGAGCAAGAAGGGTAAACAAGTTTATTTGGGGGCATATGATACTGAAGAATCTGCAGCACGTACCTATGACCT TGCAGCCCTTAAGTACTGGGGAAAAGATGCCACCCTGAATTTCCCGATAGAAACTTATACCAAGGACCTCGAGGAAATGG ACAAGGTTTCAAGAGAAGAATATTTAGCTTCTTTGCGGCGCCAAAGCAGTGGCTTTTCTAGAGGCATCTCTAAGTACCGT GGGGTTGCTAGGCATCATCATAATGGTCGCTGGGAAGCTCGCATTGGAAGAGTATGTGGAAACAAGTACCTCTACTTGGG AACATATAAAACTCAAGAGGAGGCAGCAGTGGCATATGACATGGCAGCAATTGAGTACCGTGGAGTCAATGCAGTGACCA ATTTTGACATAAGCAACTACATGGACAAAATAAAGAAGAAAAATGACCAAACCCTACAACAACAACAAACAGAAGTACAA ACAGAAACAGTTCCTAACTCCTCTGACTCTGAAGAAGCAGAAGTAGAACAACAACACACAACAACAATAACTACACCACC CCCATCTGAAAATCTGCACATGCTACCACAGGAACACCAAGTTCAATACACCCACCATGTCACTCCAAGGGATGAAGAAT CATCATCACTGGTCACAATTATGGAGCATGTGCTTGAACAGGATCTGCCATGGAGCTTCATGTACACTGGCTTGTCTCAG TTTCAAGATCCAAACTTGGCTTTAAGCAAAGGTGATGATGACTTGGCAGGCATGTTTGATGGTGCAGGGTTTGAGGAAGA CATTGATTTTCTGTTCAGCACACAACCTGGTGATCATGAGACTGAGAGTGATGTCAACAACATGAGTGCAGTTTTGGATA GTGTTGAGTGTGGAGACACAAATGGGGCTGGTGGAAGAAGCATGGTGTATCATCATGTGGATAATAATAATAAGCAGAAG AAGATGCTTTCATTTGCTTCTTCTTCTTCACCATCATCTACAACAACTACAGTTTCTTGTGACTATGCTCTAGATCTATG A |