
| Microexon ID | Gm_7:3143164-3143172:+ |
| Species | Glycine max | Coordinates | 7:3143164..3143172 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 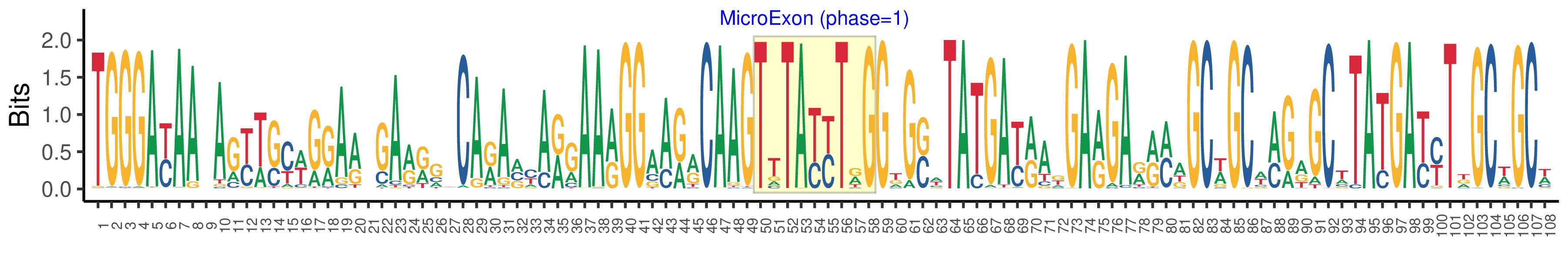 |
| Alignment of exons | 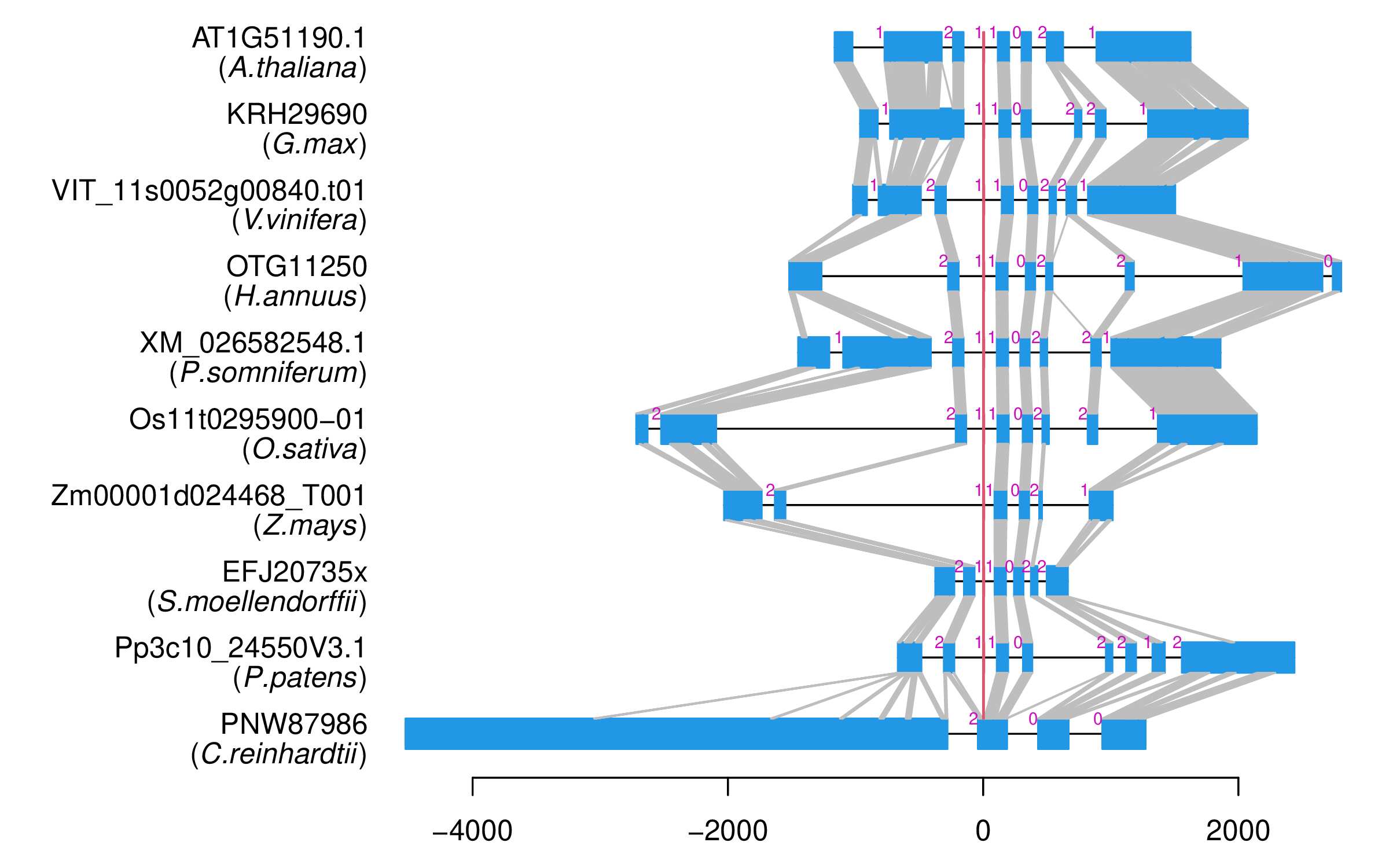 |
| Microexon DNA seq | TTTACCTTG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAATAGTTGCAGAAAGGAAGGGCAAACAAGGAAAGGAAGGCAAGTTTACCTTGGTGGTTATGATAAGGAAGAAAAAGCAGCAAAGGCTTATGACTTAGCTGCA |
| Microexon-tag Amino Acid Seq | WDNSCRKEGQTRKGRQVYLGGYDKEEKAAKAYDLAA |
| Microexon-tag spanning region | 3142883-3143426 |
| Microexon-tag prediction score | 0.9777 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH47597x |
| Reference Transcript ID | KRH47597 |
| Gene ID | GLYMA_07G038200 |
| Gene Name | NA |
| Transcript ID | KRH47597 |
| Protein ID | KRH47597 |
| Gene ID | GLYMA_07G038200 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 1.3e-12 |
| Motif start | 247 |
| Motif end | 305 |
| Protein seq | >KRH47597 MSNWLGFSLTPHLRIDEEFERENQERGAGGGGGYHHPSPSSHPHLSVMPLRSDGSLCVADSFSHSAASQEWRYDSAIGGG NSNEKGPKLEDFLGGYSNSPTKVFCQDTQHDQNQSQNNVSKINVNVAPSFCTNAEIETGESLANPYPHQSFHAYNDNSHA LIPTNGMYKSWLAQTQFSSDGKPSTEANGCNFQSLSLSMSPGMQNGVGAISSVQVSEDSRKRVMAKSHAREPVPRKSIDT FGQRTSQYRGVTRHRWTGRYEAHLWDNSCRKEGQTRKGRQVYLGGYDKEEKAAKAYDLAAIKYWGPTTHINFPLSTYEKE LEEMKHMTRQEFVANLRRKSSGFSRGASVYRGVTRHHQHGRWQARIGRVAGNKDLYLGTFSTQEEAAEAYDIAAIKFRGT SAVTNFDISRYDVKRICSSSTLIAGDLAKRSPKESPALVPAEDFNSCGSSMPPASQPPPLAITDGEHSDELSNMMWNANS DVHAQNENGANINNNVTESSSQQVSPSSNKDALNPQSPKCSVGLPNEFGVSGADYGHGYFTLDGPKYDDGNNENDHMSTN RLGNLGLVNQVPMFALWNE* |
| CDS seq | >KRH47597 ATGAGTAACTGGTTGGGGTTCTCTCTGACTCCACACTTGAGGATTGATGAAGAATTTGAGAGAGAGAACCAAGAGCGTGG AGCAGGAGGAGGAGGAGGGTACCATCATCCTTCTCCTTCTTCTCACCCTCACTTGTCTGTTATGCCTCTTCGCTCTGATG GCTCTCTCTGTGTTGCTGACTCCTTTAGCCACTCTGCTGCATCTCAAGAATGGAGATATGATAGTGCAATAGGTGGAGGG AATTCCAACGAGAAAGGTCCAAAGCTTGAGGACTTTTTGGGTGGCTACTCAAACTCCCCCACTAAGGTCTTTTGCCAAGA CACTCAGCATGACCAAAACCAAAGCCAGAACAATGTTTCCAAAATCAATGTTAATGTAGCACCAAGCTTTTGCACCAATG CAGAGATTGAAACTGGGGAGAGCCTCGCTAACCCTTATCCCCACCAATCTTTCCATGCATATAATGATAACTCACATGCC CTTATCCCCACCAATGGCATGTACAAGTCCTGGTTGGCACAAACTCAGTTTTCTTCTGATGGAAAACCTTCAACTGAAGC TAATGGGTGCAATTTCCAATCTCTGTCCCTCTCTATGAGCCCCGGTATGCAAAATGGGGTGGGTGCAATTTCCTCTGTTC AAGTGAGTGAGGATAGTAGGAAACGGGTCATGGCAAAATCTCATGCTAGAGAACCAGTCCCTCGCAAATCTATTGACACT TTTGGGCAGAGAACATCTCAATACCGTGGTGTTACAAGGCATAGATGGACTGGGAGATATGAGGCCCATTTGTGGGACAA TAGTTGCAGAAAGGAAGGGCAAACAAGGAAAGGAAGGCAAGTTTACCTTGGTGGTTATGATAAGGAAGAAAAAGCAGCAA AGGCTTATGACTTAGCTGCAATCAAGTATTGGGGTCCAACAACTCACATAAATTTCCCTTTAAGCACTTATGAGAAGGAA CTTGAAGAGATGAAGCACATGACCAGGCAAGAATTTGTCGCCAATTTAAGAAGGAAGAGCAGCGGATTTTCAAGAGGGGC ATCTGTGTATAGAGGTGTGACAAGACACCATCAACATGGAAGGTGGCAGGCCCGAATAGGAAGGGTTGCAGGAAACAAGG ACTTGTATCTTGGTACATTTAGCACACAAGAAGAAGCTGCTGAAGCATATGATATAGCAGCAATCAAGTTCAGGGGAACA AGTGCTGTGACCAACTTTGACATAAGTAGGTATGATGTGAAGAGAATCTGCTCAAGCTCCACTCTCATTGCAGGAGATCT TGCAAAACGGTCTCCTAAAGAATCACCAGCTCTGGTTCCTGCCGAGGACTTCAATTCGTGTGGTTCATCGATGCCGCCGG CATCTCAGCCCCCTCCTTTGGCTATAACTGATGGTGAACATTCTGATGAGTTGTCCAACATGATGTGGAATGCCAACAGC GATGTGCATGCACAGAATGAGAATGGTGCTAACATCAACAACAATGTCACTGAATCATCATCACAACAGGTTTCACCAAG CAGCAACAAGGATGCTTTGAACCCTCAAAGCCCCAAGTGTTCTGTGGGGCTACCCAATGAGTTTGGAGTCAGTGGAGCAG ATTATGGACATGGGTACTTCACCCTTGATGGACCAAAATATGATGATGGGAACAATGAAAACGACCACATGAGCACCAAC AGGCTTGGAAATTTGGGATTGGTTAATCAAGTTCCCATGTTTGCTTTGTGGAACGAATGA |
| Microexon DNA seq | TTTACCTTG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAATAGTTGCAGAAAGGAAGGGCAAACAAGGAAAGGAAGGCAAGTTTACCTTGGTGGTTATGATAAGGAAGAAAAAGCAGCAAAGGCTTATGACTTAGCTGCA |
| Microexon-tag Amino Acid seq | WDNSCRKEGQTRKGRQVYLGGYDKEEKAAKAYDLAA |
| Transcript ID | KRH47597 |
| Gene ID | Gm.45858 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 1.3e-12 |
| Motif start | 247 |
| Motif end | 305 |
| Protein seq | >KRH47597 MSNWLGFSLTPHLRIDEEFERENQERGAGGGGGYHHPSPSSHPHLSVMPLRSDGSLCVADSFSHSAASQEWRYDSAIGGG NSNEKGPKLEDFLGGYSNSPTKVFCQDTQHDQNQSQNNVSKINVNVAPSFCTNAEIETGESLANPYPHQSFHAYNDNSHA LIPTNGMYKSWLAQTQFSSDGKPSTEANGCNFQSLSLSMSPGMQNGVGAISSVQVSEDSRKRVMAKSHAREPVPRKSIDT FGQRTSQYRGVTRHRWTGRYEAHLWDNSCRKEGQTRKGRQVYLGGYDKEEKAAKAYDLAAIKYWGPTTHINFPLSTYEKE LEEMKHMTRQEFVANLRRKSSGFSRGASVYRGVTRHHQHGRWQARIGRVAGNKDLYLGTFSTQEEAAEAYDIAAIKFRGT SAVTNFDISRYDVKRICSSSTLIAGDLAKRSPKESPALVPAEDFNSCGSSMPPASQPPPLAITDGEHSDELSNMMWNANS DVHAQNENGANINNNVTESSSQQVSPSSNKDALNPQSPKCSVGLPNEFGVSGADYGHGYFTLDGPKYDDGNNENDHMSTN RLGNLGLVNQVPMFALWNE* |
| CDS seq | >KRH47597 ATGAGTAACTGGTTGGGGTTCTCTCTGACTCCACACTTGAGGATTGATGAAGAATTTGAGAGAGAGAACCAAGAGCGTGG AGCAGGAGGAGGAGGAGGGTACCATCATCCTTCTCCTTCTTCTCACCCTCACTTGTCTGTTATGCCTCTTCGCTCTGATG GCTCTCTCTGTGTTGCTGACTCCTTTAGCCACTCTGCTGCATCTCAAGAATGGAGATATGATAGTGCAATAGGTGGAGGG AATTCCAACGAGAAAGGTCCAAAGCTTGAGGACTTTTTGGGTGGCTACTCAAACTCCCCCACTAAGGTCTTTTGCCAAGA CACTCAGCATGACCAAAACCAAAGCCAGAACAATGTTTCCAAAATCAATGTTAATGTAGCACCAAGCTTTTGCACCAATG CAGAGATTGAAACTGGGGAGAGCCTCGCTAACCCTTATCCCCACCAATCTTTCCATGCATATAATGATAACTCACATGCC CTTATCCCCACCAATGGCATGTACAAGTCCTGGTTGGCACAAACTCAGTTTTCTTCTGATGGAAAACCTTCAACTGAAGC TAATGGGTGCAATTTCCAATCTCTGTCCCTCTCTATGAGCCCCGGTATGCAAAATGGGGTGGGTGCAATTTCCTCTGTTC AAGTGAGTGAGGATAGTAGGAAACGGGTCATGGCAAAATCTCATGCTAGAGAACCAGTCCCTCGCAAATCTATTGACACT TTTGGGCAGAGAACATCTCAATACCGTGGTGTTACAAGGCATAGATGGACTGGGAGATATGAGGCCCATTTGTGGGACAA TAGTTGCAGAAAGGAAGGGCAAACAAGGAAAGGAAGGCAAGTTTACCTTGGTGGTTATGATAAGGAAGAAAAAGCAGCAA AGGCTTATGACTTAGCTGCAATCAAGTATTGGGGTCCAACAACTCACATAAATTTCCCTTTAAGCACTTATGAGAAGGAA CTTGAAGAGATGAAGCACATGACCAGGCAAGAATTTGTCGCCAATTTAAGAAGGAAGAGCAGCGGATTTTCAAGAGGGGC ATCTGTGTATAGAGGTGTGACAAGACACCATCAACATGGAAGGTGGCAGGCCCGAATAGGAAGGGTTGCAGGAAACAAGG ACTTGTATCTTGGTACATTTAGCACACAAGAAGAAGCTGCTGAAGCATATGATATAGCAGCAATCAAGTTCAGGGGAACA AGTGCTGTGACCAACTTTGACATAAGTAGGTATGATGTGAAGAGAATCTGCTCAAGCTCCACTCTCATTGCAGGAGATCT TGCAAAACGGTCTCCTAAAGAATCACCAGCTCTGGTTCCTGCCGAGGACTTCAATTCGTGTGGTTCATCGATGCCGCCGG CATCTCAGCCCCCTCCTTTGGCTATAACTGATGGTGAACATTCTGATGAGTTGTCCAACATGATGTGGAATGCCAACAGC GATGTGCATGCACAGAATGAGAATGGTGCTAACATCAACAACAATGTCACTGAATCATCATCACAACAGGTTTCACCAAG CAGCAACAAGGATGCTTTGAACCCTCAAAGCCCCAAGTGTTCTGTGGGGCTACCCAATGAGTTTGGAGTCAGTGGAGCAG ATTATGGACATGGGTACTTCACCCTTGATGGACCAAAATATGATGATGGGAACAATGAAAACGACCACATGAGCACCAAC AGGCTTGGAAATTTGGGATTGGTTAATCAAGTTCCCATGTTTGCTTTGTGGAACGAATGA |