
| Microexon ID | Gm_18:16383135-16383143:- |
| Species | Glycine max | Coordinates | 18:16383135..16383143 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 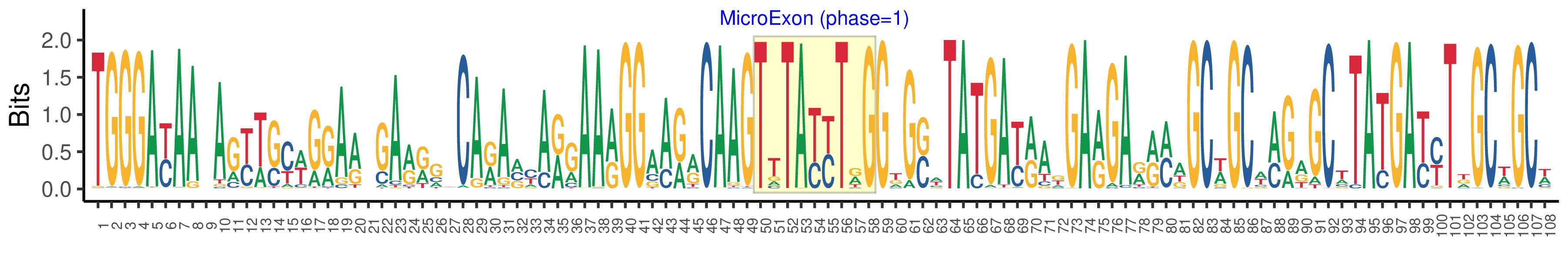 |
| Alignment of exons | 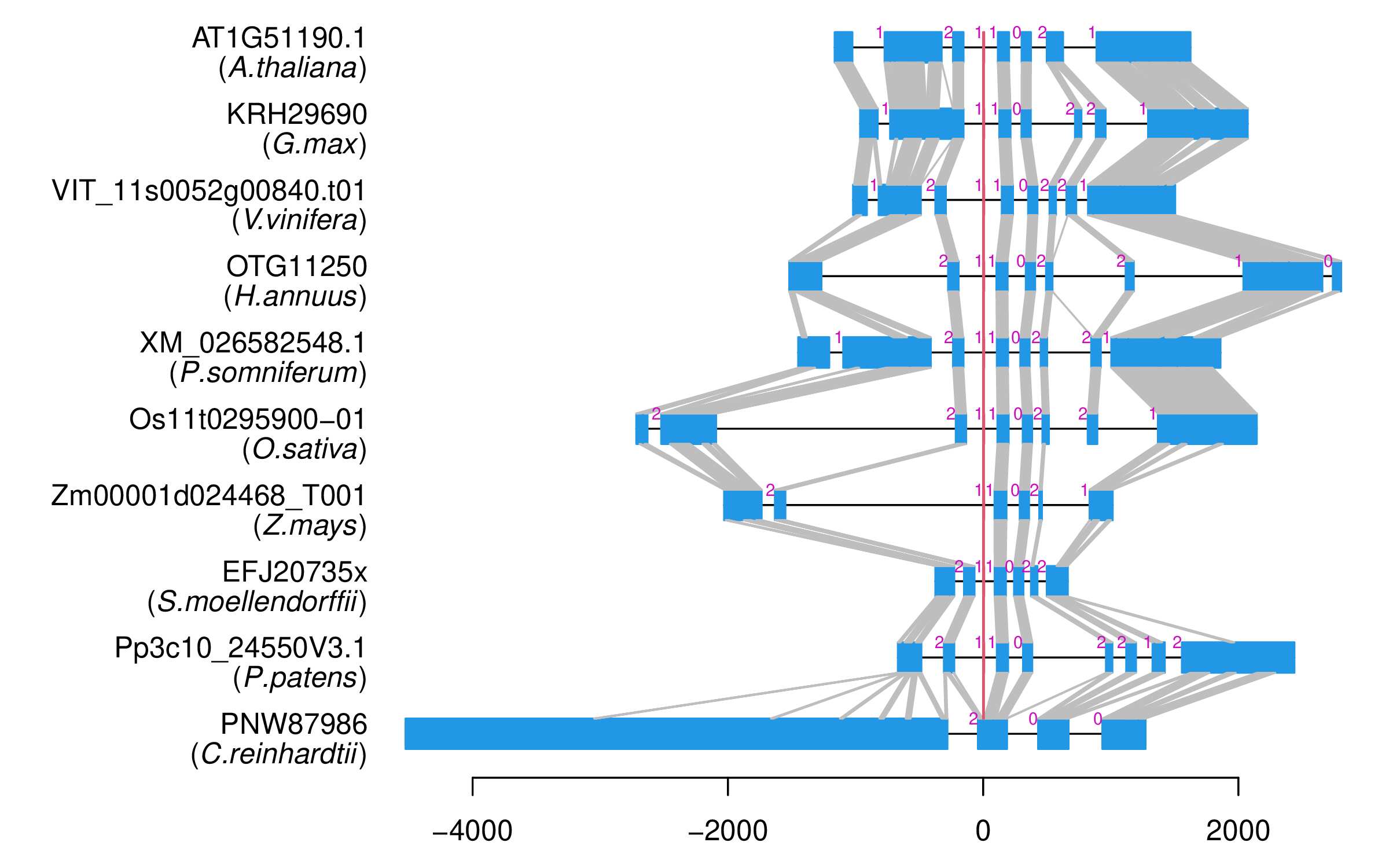 |
| Microexon DNA seq | TATATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAAAGGGACCTGGAATCCCACTCAGAAGAAGAAAGGAAAGCAAGTATATTTGGGAGCTTATAATGATGAAGAAGCTGCTGCTAGAGCTTATGATTTGGCTGCA |
| Microexon-tag Amino Acid Seq | WDKGTWNPTQKKKGKQVYLGAYNDEEAAARAYDLAA |
| Microexon-tag spanning region | 16382988-16383334 |
| Microexon-tag prediction score | 0.943 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRG99157x |
| Reference Transcript ID | KRG99157 |
| Gene ID | GLYMA_18G125200 |
| Gene Name | NA |
| Transcript ID | KRG99157 |
| Protein ID | KRG99157 |
| Gene ID | GLYMA_18G125200 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 5.6e-12 |
| Motif start | 72 |
| Motif end | 131 |
| Protein seq | >KRG99157 MELAPVKSELSPRSHRLVIIDGSDVISTKCAKRRRRDSSMAVLGGNGQQGEQLEEQKQLGGQSTATTVKRSSRFRGVSRH RWTGRFEAHLWDKGTWNPTQKKKGKQVYLGAYNDEEAAARAYDLAALKYWGTSTFTNFPVSDYEKEIEIMKTVTKEEYLA SLRRRSSGFSRGVSKYRGVARHHHNGRWEARIGRVFGNKYLYLGTYSTQEEAARAYDIAAIEYRGINAVTNFDLSTYIRW LRPGTHPTASHDQKPSTDAQLFATSNSMQTRGNIEVSNSNMHSFPSGELDSTKKRDFSKYMNPLSPCNKPSSPTALGLLL KSSVFRELMQRNLNSSSEEADVELKYPQEGNDGVGGIYDNDNTSNSYFCSSNISRLPNLESSEECPLPMYHGTMQSLWNS AFNMSN* |
| CDS seq | >KRG99157 ATGGAGCTTGCACCTGTGAAGTCTGAACTAAGTCCAAGGAGCCATAGGTTGGTCATTATAGATGGTAGTGATGTTATTAG CACTAAGTGTGCCAAAAGGCGGCGAAGAGATTCGTCTATGGCCGTGTTAGGTGGCAATGGACAACAAGGTGAACAGTTAG AAGAACAGAAGCAACTTGGTGGCCAATCAACTGCCACCACTGTGAAGAGAAGCTCAAGGTTCAGGGGAGTTAGCAGACAC AGGTGGACTGGAAGGTTTGAAGCACATCTATGGGATAAAGGGACCTGGAATCCCACTCAGAAGAAGAAAGGAAAGCAAGT ATATTTGGGAGCTTATAATGATGAAGAAGCTGCTGCTAGAGCTTATGATTTGGCTGCACTCAAGTACTGGGGAACATCAA CTTTCACAAATTTCCCTGTATCTGATTATGAGAAAGAAATTGAGATAATGAAAACTGTAACCAAAGAAGAATACCTTGCT TCATTGAGGAGGAGGAGCAGCGGTTTTTCTAGAGGTGTATCAAAGTATAGAGGAGTAGCAAGGCATCATCACAATGGAAG ATGGGAAGCAAGGATTGGGAGAGTGTTCGGCAACAAGTACCTCTACCTTGGGACTTACAGTACACAAGAAGAAGCAGCCC GTGCATATGACATTGCAGCAATTGAATACAGAGGCATAAATGCAGTGACAAATTTTGACTTGAGCACCTACATCAGATGG CTAAGACCAGGAACACATCCTACTGCTTCTCATGATCAAAAGCCTAGCACTGATGCTCAACTTTTTGCAACCTCTAACTC CATGCAAACAAGAGGGAACATTGAGGTATCCAACTCCAACATGCATTCATTCCCCTCAGGTGAATTGGACAGTACCAAGA AGCGAGACTTTTCCAAGTACATGAACCCTTTGAGTCCTTGCAACAAGCCATCTTCCCCAACAGCATTAGGACTTCTCCTA AAATCCTCGGTGTTTAGAGAACTGATGCAGAGAAATCTGAACTCTTCTAGTGAAGAAGCTGATGTTGAATTGAAATATCC ACAAGAGGGCAATGATGGGGTTGGAGGGATTTATGATAATGACAACACCAGTAACTCTTACTTTTGCTCTTCTAATATCA GCAGATTACCTAACTTGGAGTCATCAGAAGAGTGTCCATTGCCTATGTATCATGGAACTATGCAATCACTATGGAATAGT GCTTTCAACATGTCTAACTGA |
| Microexon DNA seq | TATATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAAAGGGACCTGGAATCCCACTCAGAAGAAGAAAGGAAAGCAAGTATATTTGGGAGCTTATAATGATGAAGAAGCTGCTGCTAGAGCTTATGATTTGGCTGCA |
| Microexon-tag Amino Acid seq | WDKGTWNPTQKKKGKQVYLGAYNDEEAAARAYDLAA |
| Transcript ID | KRG99157 |
| Gene ID | Gm.25136 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 5.6e-12 |
| Motif start | 72 |
| Motif end | 131 |
| Protein seq | >KRG99157 MELAPVKSELSPRSHRLVIIDGSDVISTKCAKRRRRDSSMAVLGGNGQQGEQLEEQKQLGGQSTATTVKRSSRFRGVSRH RWTGRFEAHLWDKGTWNPTQKKKGKQVYLGAYNDEEAAARAYDLAALKYWGTSTFTNFPVSDYEKEIEIMKTVTKEEYLA SLRRRSSGFSRGVSKYRGVARHHHNGRWEARIGRVFGNKYLYLGTYSTQEEAARAYDIAAIEYRGINAVTNFDLSTYIRW LRPGTHPTASHDQKPSTDAQLFATSNSMQTRGNIEVSNSNMHSFPSGELDSTKKRDFSKYMNPLSPCNKPSSPTALGLLL KSSVFRELMQRNLNSSSEEADVELKYPQEGNDGVGGIYDNDNTSNSYFCSSNISRLPNLESSEECPLPMYHGTMQSLWNS AFNMSN* |
| CDS seq | >KRG99157 ATGGAGCTTGCACCTGTGAAGTCTGAACTAAGTCCAAGGAGCCATAGGTTGGTCATTATAGATGGTAGTGATGTTATTAG CACTAAGTGTGCCAAAAGGCGGCGAAGAGATTCGTCTATGGCCGTGTTAGGTGGCAATGGACAACAAGGTGAACAGTTAG AAGAACAGAAGCAACTTGGTGGCCAATCAACTGCCACCACTGTGAAGAGAAGCTCAAGGTTCAGGGGAGTTAGCAGACAC AGGTGGACTGGAAGGTTTGAAGCACATCTATGGGATAAAGGGACCTGGAATCCCACTCAGAAGAAGAAAGGAAAGCAAGT ATATTTGGGAGCTTATAATGATGAAGAAGCTGCTGCTAGAGCTTATGATTTGGCTGCACTCAAGTACTGGGGAACATCAA CTTTCACAAATTTCCCTGTATCTGATTATGAGAAAGAAATTGAGATAATGAAAACTGTAACCAAAGAAGAATACCTTGCT TCATTGAGGAGGAGGAGCAGCGGTTTTTCTAGAGGTGTATCAAAGTATAGAGGAGTAGCAAGGCATCATCACAATGGAAG ATGGGAAGCAAGGATTGGGAGAGTGTTCGGCAACAAGTACCTCTACCTTGGGACTTACAGTACACAAGAAGAAGCAGCCC GTGCATATGACATTGCAGCAATTGAATACAGAGGCATAAATGCAGTGACAAATTTTGACTTGAGCACCTACATCAGATGG CTAAGACCAGGAACACATCCTACTGCTTCTCATGATCAAAAGCCTAGCACTGATGCTCAACTTTTTGCAACCTCTAACTC CATGCAAACAAGAGGGAACATTGAGGTATCCAACTCCAACATGCATTCATTCCCCTCAGGTGAATTGGACAGTACCAAGA AGCGAGACTTTTCCAAGTACATGAACCCTTTGAGTCCTTGCAACAAGCCATCTTCCCCAACAGCATTAGGACTTCTCCTA AAATCCTCGGTGTTTAGAGAACTGATGCAGAGAAATCTGAACTCTTCTAGTGAAGAAGCTGATGTTGAATTGAAATATCC ACAAGAGGGCAATGATGGGGTTGGAGGGATTTATGATAATGACAACACCAGTAACTCTTACTTTTGCTCTTCTAATATCA GCAGATTACCTAACTTGGAGTCATCAGAAGAGTGTCCATTGCCTATGTATCATGGAACTATGCAATCACTATGGAATAGT GCTTTCAACATGTCTAACTGA |