
| Microexon ID | Gm_17:5548302-5548310:- |
| Species | Glycine max | Coordinates | 17:5548302..5548310 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 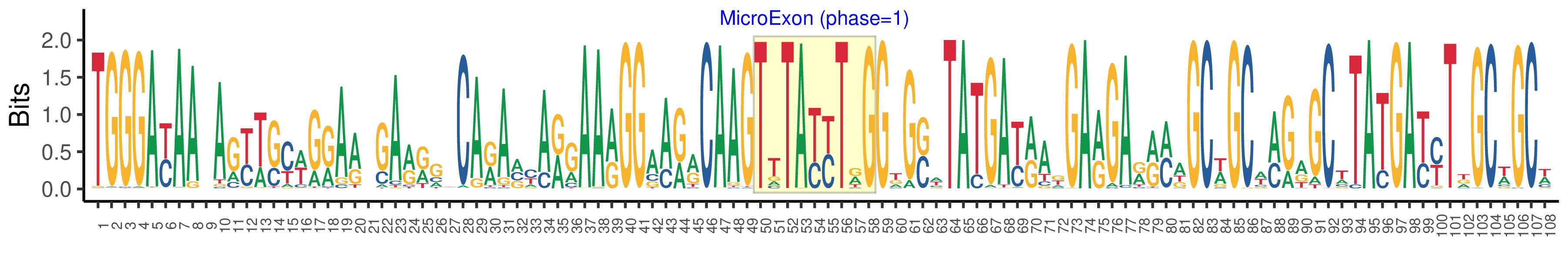 |
| Alignment of exons | 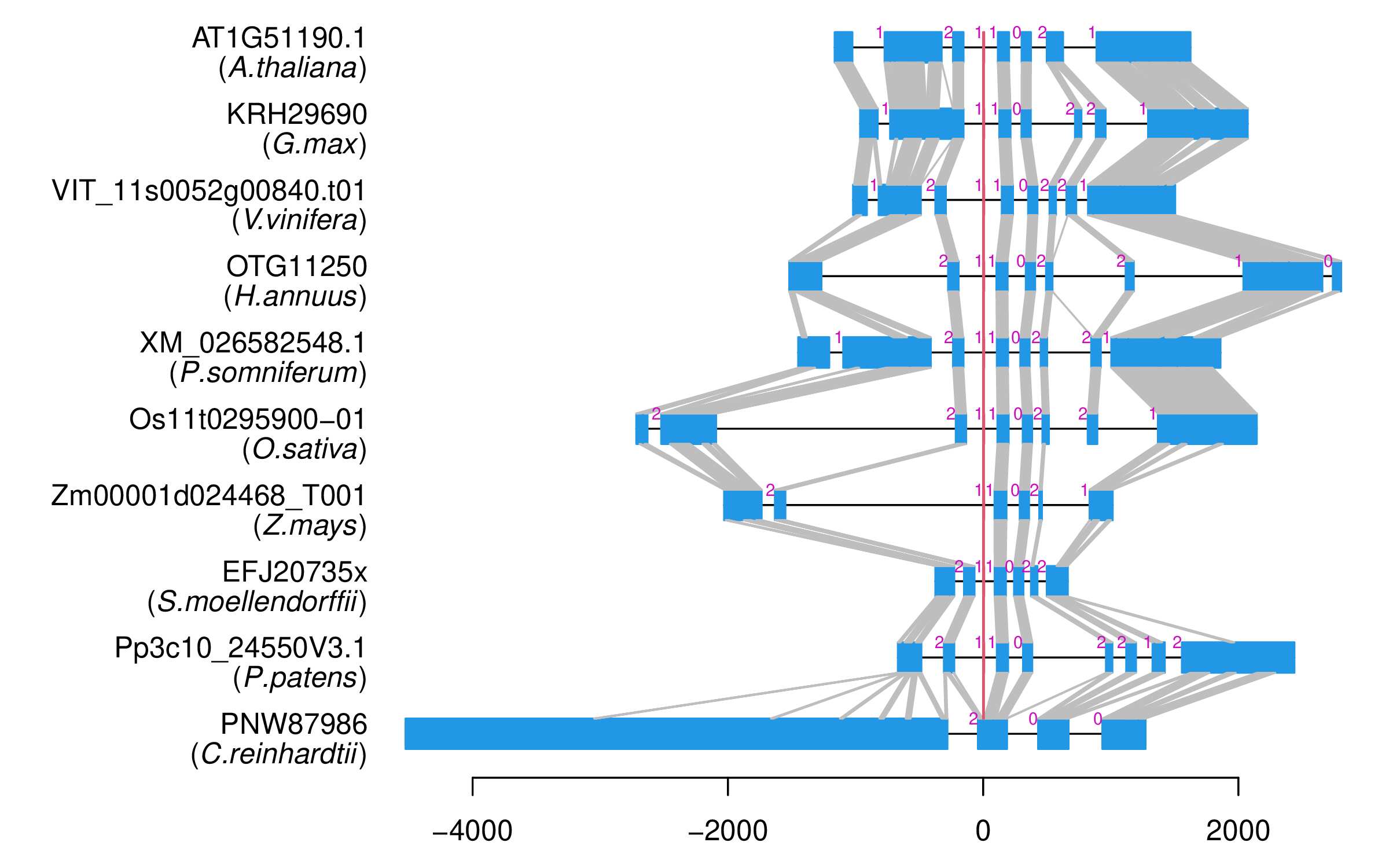 |
| Microexon DNA seq | TCTACCTTG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAAACATTGCTGGAATGAATCACAGAACAAAAAAGGGCGACAAGTCTACCTTGGCGCTTATGACAATGAAGAGGCAGCAGCACATGCTTATGATCTAGCAGCA |
| Microexon-tag Amino Acid Seq | WDKHCWNESQNKKGRQVYLGAYDNEEAAAHAYDLAA |
| Microexon-tag spanning region | 5548165-5548514 |
| Microexon-tag prediction score | 0.9576 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH02994x |
| Reference Transcript ID | KRH02994 |
| Gene ID | GLYMA_17G070800 |
| Gene Name | NA |
| Transcript ID | KRH02994 |
| Protein ID | KRH02994 |
| Gene ID | GLYMA_17G070800 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 4.6e-13 |
| Motif start | 50 |
| Motif end | 108 |
| Protein seq | >KRH02994 MAKKSQLRTQKNNVTTNDDNNLNVTNTVTTKVKRTRRSVPRDSPPQRSSIYRGVTRHRWTGRYEAHLWDKHCWNESQNKK GRQVYLGAYDNEEAAAHAYDLAALKYWGQDTILNFPLSNYLNELKEMEGQSREEYIGSLRRKSSGFSRGISKYRGVARHH HNGRWEARIGKVFGNKYLYLGTYATQEEAATAYDLAAIEYRGLNAVTNFDLSRYIKWLKPNNNTNNVIDDQISINLTNIN NNNNCTNSFTPSPDQEQEASFFHNKDSLNNTIVEEVTLVPHQPRPASATSALELLLQSSKFKEMMEMTSVANLSSTQMES ELPQCTFPDHIQTYFEYEDSNRYEEGDDLMFKFNEFSSIVPFYQCDEFES* |
| CDS seq | >KRH02994 ATGGCCAAAAAATCACAGCTGCGTACCCAGAAAAACAATGTTACCACCAATGACGATAATAATCTTAACGTAACCAACAC TGTGACCACCAAGGTGAAACGAACAAGGAGAAGTGTCCCTAGAGACTCCCCACCTCAACGCAGCTCAATATACCGAGGAG TCACTAGGCACCGATGGACAGGCCGATACGAAGCTCATTTGTGGGACAAACATTGCTGGAATGAATCACAGAACAAAAAA GGGCGACAAGTCTACCTTGGCGCTTATGACAATGAAGAGGCAGCAGCACATGCTTATGATCTAGCAGCACTGAAATACTG GGGTCAAGATACCATTCTTAATTTTCCGTTATCAAACTACCTGAACGAACTGAAAGAAATGGAGGGTCAATCACGGGAGG AGTATATCGGATCGCTGAGGAGGAAAAGCAGTGGTTTTTCTCGGGGAATTTCTAAATACAGAGGTGTTGCAAGGCATCAT CATAACGGAAGGTGGGAGGCTCGGATTGGCAAAGTTTTTGGCAATAAATATCTTTACCTCGGAACTTATGCTACCCAAGA AGAAGCTGCTACTGCCTATGACCTGGCAGCCATAGAATACCGTGGACTCAATGCTGTCACCAATTTCGATCTCAGCCGTT ACATTAAGTGGCTTAAGCCTAACAACAACACCAACAACGTTATCGACGACCAGATTAGTATTAATCTCACTAACATAAAC AATAATAATAATTGCACTAACAGCTTCACCCCAAGTCCTGATCAAGAACAAGAAGCTAGCTTCTTCCACAACAAAGATTC ACTCAATAATACTATTGTAGAAGAAGTCACGTTGGTGCCACATCAGCCTCGTCCAGCGAGTGCCACGTCAGCATTGGAGC TTCTACTTCAGTCATCAAAGTTCAAGGAAATGATGGAGATGACATCTGTGGCCAATCTTTCATCAACACAGATGGAATCT GAGTTGCCACAGTGCACATTTCCTGATCACATTCAGACGTACTTTGAGTATGAAGATTCCAATAGATATGAGGAAGGAGA TGATCTCATGTTCAAGTTCAACGAGTTCAGCTCCATTGTGCCGTTTTACCAATGTGACGAGTTCGAGAGTTGA |
| Microexon DNA seq | TCTACCTTG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAAACATTGCTGGAATGAATCACAGAACAAAAAAGGGCGACAAGTCTACCTTGGCGCTTATGACAATGAAGAGGCAGCAGCACATGCTTATGATCTAGCAGCA |
| Microexon-tag Amino Acid seq | WDKHCWNESQNKKGRQVYLGAYDNEEAAAHAYDLAA |
| Transcript ID | KRH02994 |
| Gene ID | Gm.22097 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 4.6e-13 |
| Motif start | 50 |
| Motif end | 108 |
| Protein seq | >KRH02994 MAKKSQLRTQKNNVTTNDDNNLNVTNTVTTKVKRTRRSVPRDSPPQRSSIYRGVTRHRWTGRYEAHLWDKHCWNESQNKK GRQVYLGAYDNEEAAAHAYDLAALKYWGQDTILNFPLSNYLNELKEMEGQSREEYIGSLRRKSSGFSRGISKYRGVARHH HNGRWEARIGKVFGNKYLYLGTYATQEEAATAYDLAAIEYRGLNAVTNFDLSRYIKWLKPNNNTNNVIDDQISINLTNIN NNNNCTNSFTPSPDQEQEASFFHNKDSLNNTIVEEVTLVPHQPRPASATSALELLLQSSKFKEMMEMTSVANLSSTQMES ELPQCTFPDHIQTYFEYEDSNRYEEGDDLMFKFNEFSSIVPFYQCDEFES* |
| CDS seq | >KRH02994 ATGGCCAAAAAATCACAGCTGCGTACCCAGAAAAACAATGTTACCACCAATGACGATAATAATCTTAACGTAACCAACAC TGTGACCACCAAGGTGAAACGAACAAGGAGAAGTGTCCCTAGAGACTCCCCACCTCAACGCAGCTCAATATACCGAGGAG TCACTAGGCACCGATGGACAGGCCGATACGAAGCTCATTTGTGGGACAAACATTGCTGGAATGAATCACAGAACAAAAAA GGGCGACAAGTCTACCTTGGCGCTTATGACAATGAAGAGGCAGCAGCACATGCTTATGATCTAGCAGCACTGAAATACTG GGGTCAAGATACCATTCTTAATTTTCCGTTATCAAACTACCTGAACGAACTGAAAGAAATGGAGGGTCAATCACGGGAGG AGTATATCGGATCGCTGAGGAGGAAAAGCAGTGGTTTTTCTCGGGGAATTTCTAAATACAGAGGTGTTGCAAGGCATCAT CATAACGGAAGGTGGGAGGCTCGGATTGGCAAAGTTTTTGGCAATAAATATCTTTACCTCGGAACTTATGCTACCCAAGA AGAAGCTGCTACTGCCTATGACCTGGCAGCCATAGAATACCGTGGACTCAATGCTGTCACCAATTTCGATCTCAGCCGTT ACATTAAGTGGCTTAAGCCTAACAACAACACCAACAACGTTATCGACGACCAGATTAGTATTAATCTCACTAACATAAAC AATAATAATAATTGCACTAACAGCTTCACCCCAAGTCCTGATCAAGAACAAGAAGCTAGCTTCTTCCACAACAAAGATTC ACTCAATAATACTATTGTAGAAGAAGTCACGTTGGTGCCACATCAGCCTCGTCCAGCGAGTGCCACGTCAGCATTGGAGC TTCTACTTCAGTCATCAAAGTTCAAGGAAATGATGGAGATGACATCTGTGGCCAATCTTTCATCAACACAGATGGAATCT GAGTTGCCACAGTGCACATTTCCTGATCACATTCAGACGTACTTTGAGTATGAAGATTCCAATAGATATGAGGAAGGAGA TGATCTCATGTTCAAGTTCAACGAGTTCAGCTCCATTGTGCCGTTTTACCAATGTGACGAGTTCGAGAGTTGA |