
| Microexon ID | Gm_16:607211-607219:+ |
| Species | Glycine max | Coordinates | 16:607211..607219 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 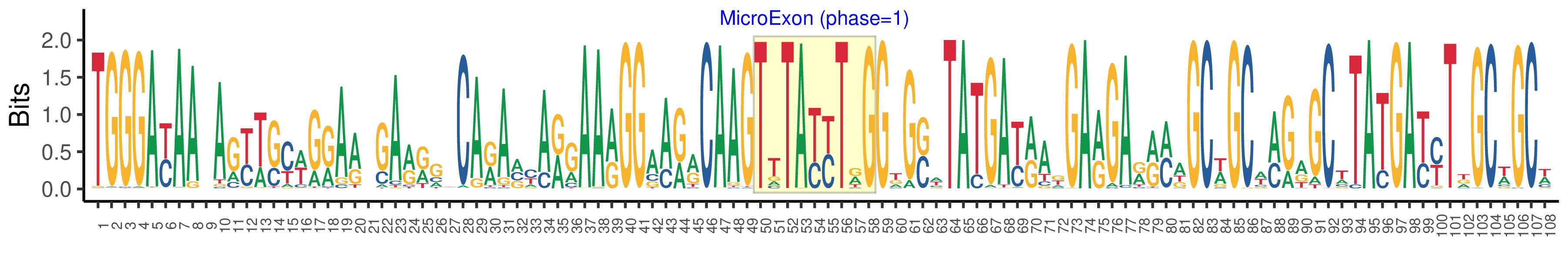 |
| Alignment of exons | 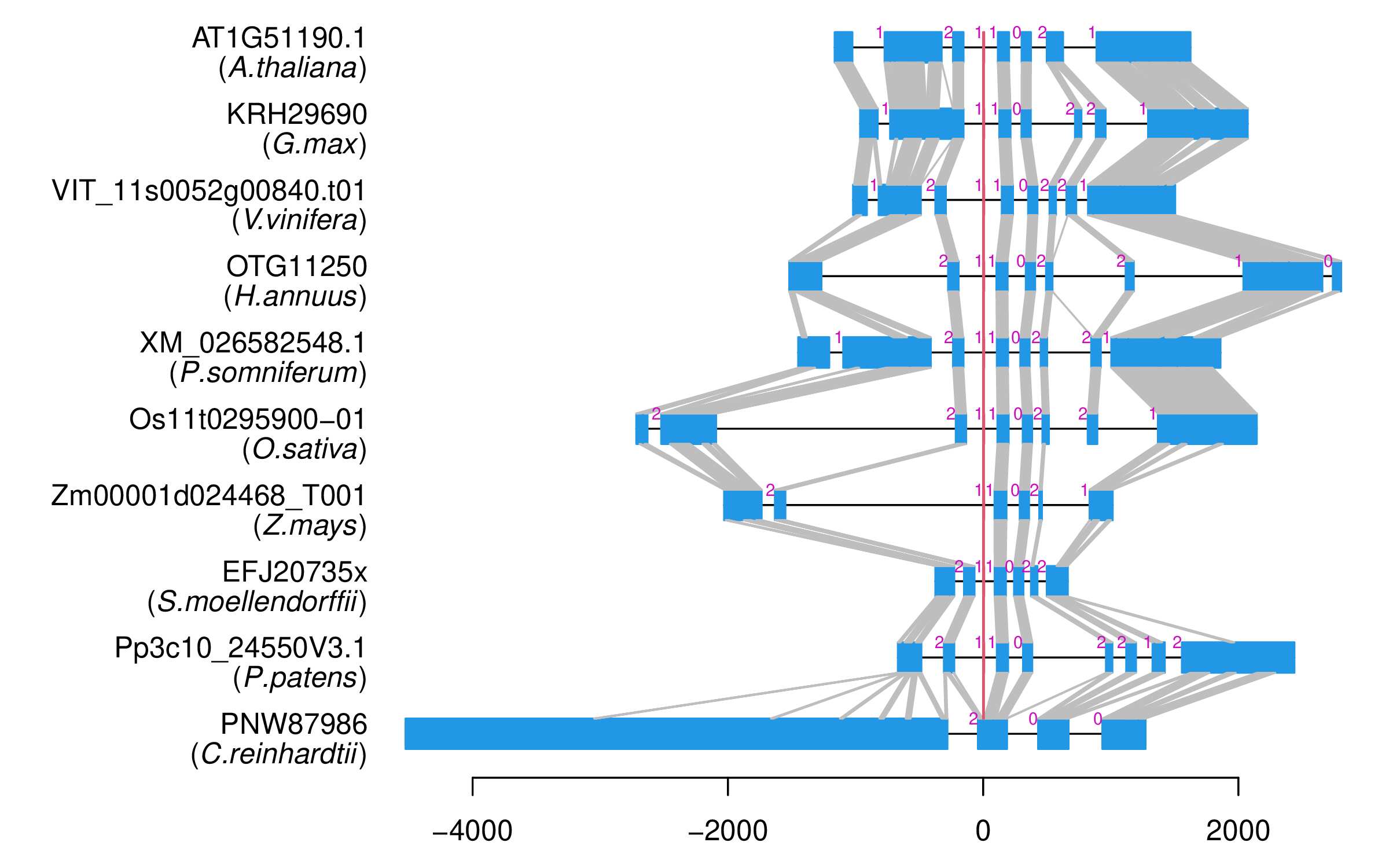 |
| Microexon DNA seq | TTTACCTTG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAATAGTTGCAGAAAGGAAGGGCAAACAAGGAAAGGAAGGCAAGTTTACCTTGGTGGTTATGATAAGGAAGAAAAAGCAGCAAAGGCTTACGACTTAGCTGCA |
| Microexon-tag Amino Acid Seq | WDNSCRKEGQTRKGRQVYLGGYDKEEKAAKAYDLAA |
| Microexon-tag spanning region | 606950-607476 |
| Microexon-tag prediction score | 0.9756 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH06179x |
| Reference Transcript ID | KRH06179 |
| Gene ID | GLYMA_16G007400 |
| Gene Name | NA |
| Transcript ID | KRH06179 |
| Protein ID | KRH06179 |
| Gene ID | GLYMA_16G007400 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 6.9e-13 |
| Motif start | 245 |
| Motif end | 303 |
| Protein seq | >KRH06179 MSNWLGFSLTPHLRIDEEFERENQERGGGYHHPSPSCHPHLSVMPLRSDGSLCVADSFSHSAASQEWRYDSAIGGGNSNE EGPKLEDFLGCYSNSPAKVFCQDSQPDQNQSQNNVSKINVNVAPSFCTNNSEIETGDNLTNPSSLLHSFHAYNDNSHALI PTNGMYKSWLAQTQFSSDGKPSNEANGCNFQSLSLTMSPSVQNGVGAISSVQVNEDSRKRVMAKSHAREPVPRKSIDTFG QRTSQYRGVTRHRWTGRYEAHLWDNSCRKEGQTRKGRQVYLGGYDKEEKAAKAYDLAALKYWGPTTHINFPLSTYEKELE EMKHMTRQEFVANLRRKSSGFSRGASVYRGVTRHHQHGRWQARIGRVAGNKDLYLGTFSTQEEAAEAYDIAAIKFRGTSA VTNFDISRYDVKRICSSSTLIAGDLAKRSPKESPAPVPASDFNSCGSSPMPLVSQPPPLAITDGEHSDELSNMMWNANNS DEQAQNESGGAEFNNNVTESSSSQQVSPSSNKDALNPQSPNEFGVSGADYGHGYFTLDGPKYDDGNNENDHMSTNRLGNL GLVNQVPMFALWNE* |
| CDS seq | >KRH06179 ATGAGTAACTGGTTGGGGTTCTCTCTCACTCCACACTTGAGGATTGATGAAGAATTTGAGAGAGAAAACCAAGAGCGTGG AGGAGGGTACCATCATCCTTCTCCTTCTTGTCACCCTCACTTGTCTGTTATGCCCCTTCGCTCTGATGGCTCTCTCTGTG TTGCTGACTCCTTTAGTCACTCTGCTGCATCTCAAGAATGGAGATATGATAGTGCAATAGGTGGAGGGAATTCCAATGAA GAAGGTCCAAAGCTTGAAGACTTTTTGGGTTGCTACTCAAACTCCCCCGCTAAGGTCTTTTGCCAAGACTCTCAGCCTGA CCAAAACCAAAGCCAGAACAATGTTTCCAAAATCAATGTTAATGTGGCACCAAGCTTTTGCACCAACAATTCAGAGATTG AAACTGGGGATAATCTCACAAATCCTTCTTCCTTGCTCCATTCTTTCCATGCATATAATGATAACTCACATGCTCTTATC CCCACCAATGGCATGTACAAGTCCTGGTTGGCACAAACCCAGTTTTCTTCTGATGGAAAACCTTCAAATGAGGCTAATGG GTGCAATTTCCAATCTTTGTCCCTCACTATGAGCCCCAGTGTGCAAAATGGGGTGGGTGCAATTTCCTCTGTTCAAGTGA ATGAGGATAGTAGGAAACGGGTCATGGCGAAATCTCATGCTAGAGAACCAGTTCCTCGCAAATCTATTGACACTTTTGGG CAAAGAACATCTCAATATCGTGGTGTTACAAGGCATAGATGGACTGGAAGATATGAGGCCCATTTGTGGGATAATAGTTG CAGAAAGGAAGGGCAAACAAGGAAAGGAAGGCAAGTTTACCTTGGTGGTTATGATAAGGAAGAAAAAGCAGCAAAGGCTT ACGACTTAGCTGCACTCAAGTATTGGGGTCCAACAACTCACATAAATTTCCCTTTAAGCACTTATGAGAAGGAACTTGAA GAGATGAAGCACATGACCAGGCAAGAATTTGTCGCCAATTTAAGAAGGAAGAGCAGTGGATTTTCAAGAGGGGCATCTGT GTATAGAGGTGTGACAAGACACCATCAACATGGAAGGTGGCAGGCCCGAATAGGAAGGGTTGCAGGAAACAAGGACTTGT ATCTTGGTACATTTAGCACACAAGAAGAAGCTGCTGAAGCATATGATATAGCAGCAATCAAGTTCAGAGGAACAAGTGCT GTGACCAACTTTGACATAAGTAGGTATGATGTGAAGAGAATCTGCTCAAGCTCCACTCTTATTGCAGGAGATCTTGCAAA ACGGTCTCCTAAAGAATCACCAGCTCCGGTTCCTGCCTCTGACTTCAATTCGTGCGGTTCATCGCCGATGCCGCTGGTGT CTCAGCCACCTCCTTTGGCTATAACTGATGGTGAACATTCTGATGAGTTGTCCAACATGATGTGGAATGCCAACAACAGT GATGAGCAAGCACAGAATGAGAGTGGTGGTGCTGAATTCAACAACAATGTAACTGAATCATCATCATCACAACAGGTTTC ACCAAGCAGCAACAAGGATGCTTTGAACCCTCAAAGCCCCAATGAGTTTGGAGTCAGTGGAGCAGATTATGGACATGGCT ACTTCACCCTTGATGGACCAAAATATGATGATGGGAACAATGAAAATGACCACATGAGCACCAACAGGCTTGGAAATTTG GGATTGGTTAATCAAGTTCCCATGTTTGCTTTGTGGAATGAATGA |
| Microexon DNA seq | TTTACCTTG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAATAGTTGCAGAAAGGAAGGGCAAACAAGGAAAGGAAGGCAAGTTTACCTTGGTGGTTATGATAAGGAAGAAAAAGCAGCAAAGGCTTACGACTTAGCTGCA |
| Microexon-tag Amino Acid seq | WDNSCRKEGQTRKGRQVYLGGYDKEEKAAKAYDLAA |
| Transcript ID | KRH06179 |
| Gene ID | Gm.19321 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 6.9e-13 |
| Motif start | 245 |
| Motif end | 303 |
| Protein seq | >KRH06179 MSNWLGFSLTPHLRIDEEFERENQERGGGYHHPSPSCHPHLSVMPLRSDGSLCVADSFSHSAASQEWRYDSAIGGGNSNE EGPKLEDFLGCYSNSPAKVFCQDSQPDQNQSQNNVSKINVNVAPSFCTNNSEIETGDNLTNPSSLLHSFHAYNDNSHALI PTNGMYKSWLAQTQFSSDGKPSNEANGCNFQSLSLTMSPSVQNGVGAISSVQVNEDSRKRVMAKSHAREPVPRKSIDTFG QRTSQYRGVTRHRWTGRYEAHLWDNSCRKEGQTRKGRQVYLGGYDKEEKAAKAYDLAALKYWGPTTHINFPLSTYEKELE EMKHMTRQEFVANLRRKSSGFSRGASVYRGVTRHHQHGRWQARIGRVAGNKDLYLGTFSTQEEAAEAYDIAAIKFRGTSA VTNFDISRYDVKRICSSSTLIAGDLAKRSPKESPAPVPASDFNSCGSSPMPLVSQPPPLAITDGEHSDELSNMMWNANNS DEQAQNESGGAEFNNNVTESSSSQQVSPSSNKDALNPQSPNEFGVSGADYGHGYFTLDGPKYDDGNNENDHMSTNRLGNL GLVNQVPMFALWNE* |
| CDS seq | >KRH06179 ATGAGTAACTGGTTGGGGTTCTCTCTCACTCCACACTTGAGGATTGATGAAGAATTTGAGAGAGAAAACCAAGAGCGTGG AGGAGGGTACCATCATCCTTCTCCTTCTTGTCACCCTCACTTGTCTGTTATGCCCCTTCGCTCTGATGGCTCTCTCTGTG TTGCTGACTCCTTTAGTCACTCTGCTGCATCTCAAGAATGGAGATATGATAGTGCAATAGGTGGAGGGAATTCCAATGAA GAAGGTCCAAAGCTTGAAGACTTTTTGGGTTGCTACTCAAACTCCCCCGCTAAGGTCTTTTGCCAAGACTCTCAGCCTGA CCAAAACCAAAGCCAGAACAATGTTTCCAAAATCAATGTTAATGTGGCACCAAGCTTTTGCACCAACAATTCAGAGATTG AAACTGGGGATAATCTCACAAATCCTTCTTCCTTGCTCCATTCTTTCCATGCATATAATGATAACTCACATGCTCTTATC CCCACCAATGGCATGTACAAGTCCTGGTTGGCACAAACCCAGTTTTCTTCTGATGGAAAACCTTCAAATGAGGCTAATGG GTGCAATTTCCAATCTTTGTCCCTCACTATGAGCCCCAGTGTGCAAAATGGGGTGGGTGCAATTTCCTCTGTTCAAGTGA ATGAGGATAGTAGGAAACGGGTCATGGCGAAATCTCATGCTAGAGAACCAGTTCCTCGCAAATCTATTGACACTTTTGGG CAAAGAACATCTCAATATCGTGGTGTTACAAGGCATAGATGGACTGGAAGATATGAGGCCCATTTGTGGGATAATAGTTG CAGAAAGGAAGGGCAAACAAGGAAAGGAAGGCAAGTTTACCTTGGTGGTTATGATAAGGAAGAAAAAGCAGCAAAGGCTT ACGACTTAGCTGCACTCAAGTATTGGGGTCCAACAACTCACATAAATTTCCCTTTAAGCACTTATGAGAAGGAACTTGAA GAGATGAAGCACATGACCAGGCAAGAATTTGTCGCCAATTTAAGAAGGAAGAGCAGTGGATTTTCAAGAGGGGCATCTGT GTATAGAGGTGTGACAAGACACCATCAACATGGAAGGTGGCAGGCCCGAATAGGAAGGGTTGCAGGAAACAAGGACTTGT ATCTTGGTACATTTAGCACACAAGAAGAAGCTGCTGAAGCATATGATATAGCAGCAATCAAGTTCAGAGGAACAAGTGCT GTGACCAACTTTGACATAAGTAGGTATGATGTGAAGAGAATCTGCTCAAGCTCCACTCTTATTGCAGGAGATCTTGCAAA ACGGTCTCCTAAAGAATCACCAGCTCCGGTTCCTGCCTCTGACTTCAATTCGTGCGGTTCATCGCCGATGCCGCTGGTGT CTCAGCCACCTCCTTTGGCTATAACTGATGGTGAACATTCTGATGAGTTGTCCAACATGATGTGGAATGCCAACAACAGT GATGAGCAAGCACAGAATGAGAGTGGTGGTGCTGAATTCAACAACAATGTAACTGAATCATCATCATCACAACAGGTTTC ACCAAGCAGCAACAAGGATGCTTTGAACCCTCAAAGCCCCAATGAGTTTGGAGTCAGTGGAGCAGATTATGGACATGGCT ACTTCACCCTTGATGGACCAAAATATGATGATGGGAACAATGAAAATGACCACATGAGCACCAACAGGCTTGGAAATTTG GGATTGGTTAATCAAGTTCCCATGTTTGCTTTGTGGAATGAATGA |