
| Microexon ID | Gm_15:40068827-40068835:+ |
| Species | Glycine max | Coordinates | 15:40068827..40068835 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 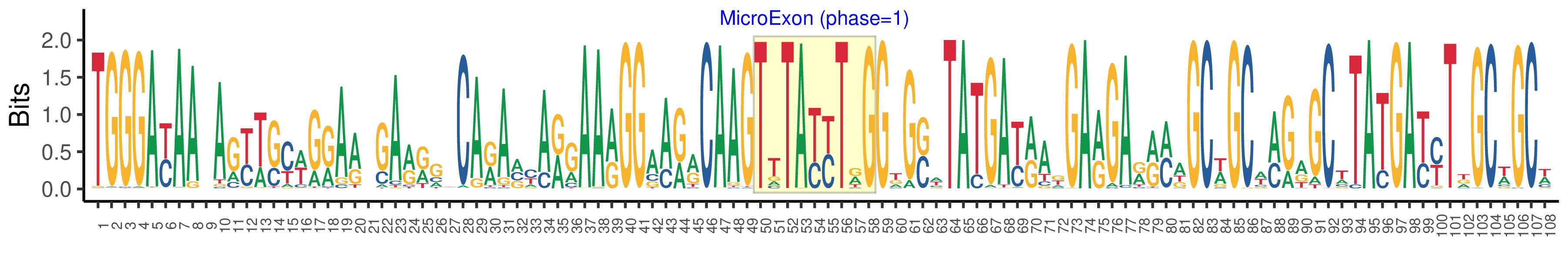 |
| Alignment of exons | 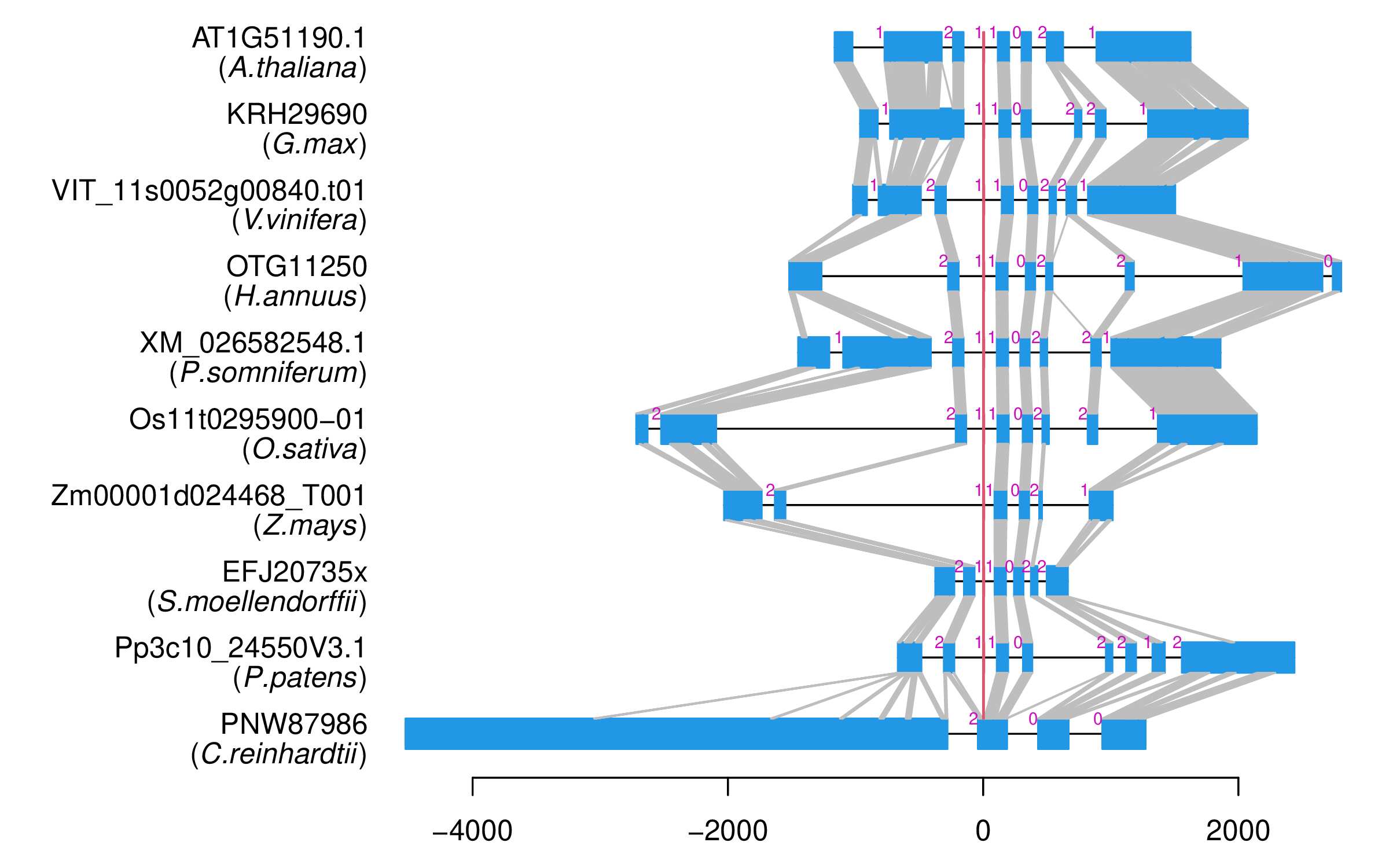 |
| Microexon DNA seq | TTTATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAAGAGCTCTTGGAACAACATTCAGAGCAAGAAGGGTCGACAAGTTTATTTGGGGGCATATGATACTGAAGAATCTGCAGCCCGTACCTATGACCTTGCAGCC |
| Microexon-tag Amino Acid Seq | WDKSSWNNIQSKKGRQVYLGAYDTEESAARTYDLAA |
| Microexon-tag spanning region | 40068565-40069072 |
| Microexon-tag prediction score | 0.9447 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH13193x |
| Reference Transcript ID | KRH13193 |
| Gene ID | GLYMA_15G221600 |
| Gene Name | NA |
| Transcript ID | KRH13193 |
| Protein ID | KRH13193 |
| Gene ID | GLYMA_15G221600 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 3.5e-13 |
| Motif start | 54 |
| Motif end | 112 |
| Protein seq | >KRH13193 MKRSPASSCSSSTSSVGFEAPIEKRRPKHPRRNNLKSQKCKQNQTTTGGRRSSIYRGVTRHRWTGRFEAHLWDKSSWNNI QSKKGRQVYLGAYDTEESAARTYDLAALKYWGKDATLNFPIETYTKELEEMDKVSREEYLASLRRQSSGFSRGLSKYRGV ARHHHNGRWEARIGRVCGNKYLYLGTYKTQEEAAVAYDMAAIEYRGVNAVTNFDISNYMDKIKKKNDQTQQQQTEAQTET VPNSSDSEEVEVEQQTTTITTPPPSENLHMPPQQHQVQYTPHVSPREEESSSLITIMDHVLEQDLPWSFMYTGLSQFQDP NLAFCKGDDDLVGMFDSAGFEEDIDFLFSTQPGDETESDVNNMSAVLDSVECGDTNGAGGSMMHVDNKQKIVSFASSPSS TTTVSCDYALDL* |
| CDS seq | >KRH13193 ATGAAGAGGTCTCCAGCATCTTCTTGTTCATCATCTACTTCCTCTGTTGGGTTTGAAGCTCCCATTGAAAAAAGAAGGCC TAAGCATCCAAGGAGGAATAATTTGAAGTCACAAAAATGCAAGCAGAACCAAACCACCACTGGTGGCAGAAGAAGCTCTA TCTATAGAGGAGTTACAAGGCATAGGTGGACAGGGAGGTTTGAAGCTCACCTATGGGATAAGAGCTCTTGGAACAACATT CAGAGCAAGAAGGGTCGACAAGTTTATTTGGGGGCATATGATACTGAAGAATCTGCAGCCCGTACCTATGACCTTGCAGC CCTTAAATACTGGGGAAAAGATGCAACCCTGAATTTCCCGATAGAAACTTATACCAAGGAGCTCGAGGAAATGGACAAGG TTTCAAGAGAAGAATATTTGGCTTCTTTGCGGCGCCAAAGCAGTGGCTTTTCTAGAGGCCTGTCTAAGTACCGTGGGGTT GCTAGGCATCATCATAATGGTCGCTGGGAAGCACGAATTGGAAGAGTATGCGGAAACAAGTACCTCTACTTGGGGACATA TAAAACTCAAGAGGAGGCAGCAGTGGCATATGACATGGCAGCAATAGAGTACCGTGGAGTCAATGCAGTGACCAATTTTG ACATAAGCAACTACATGGACAAAATAAAGAAGAAAAATGACCAAACCCAACAACAACAAACAGAAGCACAAACGGAAACA GTTCCTAACTCCTCTGACTCTGAAGAAGTAGAAGTAGAACAACAGACAACAACAATAACCACACCACCCCCATCTGAAAA TCTGCACATGCCACCACAGCAGCACCAAGTTCAATACACCCCCCATGTCTCTCCAAGGGAAGAAGAATCATCATCACTGA TCACAATTATGGACCATGTGCTTGAGCAGGATCTGCCATGGAGCTTCATGTACACTGGCTTGTCTCAGTTTCAAGATCCA AACTTGGCTTTCTGCAAAGGTGATGATGACTTGGTGGGCATGTTTGATAGTGCAGGGTTTGAGGAAGACATTGATTTTCT GTTCAGCACTCAACCTGGTGATGAGACTGAGAGTGATGTCAACAATATGAGCGCAGTTTTGGATAGTGTTGAGTGTGGAG ACACAAATGGGGCTGGTGGAAGCATGATGCATGTGGATAACAAGCAGAAGATAGTATCATTTGCTTCTTCACCATCATCT ACAACTACAGTTTCTTGTGACTATGCTCTAGATCTATGA |
| Microexon DNA seq | TTTATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAAGAGCTCTTGGAACAACATTCAGAGCAAGAAGGGTCGACAAGTTTATTTGGGGGCATATGATACTGAAGAATCTGCAGCCCGTACCTATGACCTTGCAGCC |
| Microexon-tag Amino Acid seq | WDKSSWNNIQSKKGRQVYLGAYDTEESAARTYDLAA |
| Transcript ID | KRH13193 |
| Gene ID | Gm.18703 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 3.5e-13 |
| Motif start | 54 |
| Motif end | 112 |
| Protein seq | >KRH13193 MKRSPASSCSSSTSSVGFEAPIEKRRPKHPRRNNLKSQKCKQNQTTTGGRRSSIYRGVTRHRWTGRFEAHLWDKSSWNNI QSKKGRQVYLGAYDTEESAARTYDLAALKYWGKDATLNFPIETYTKELEEMDKVSREEYLASLRRQSSGFSRGLSKYRGV ARHHHNGRWEARIGRVCGNKYLYLGTYKTQEEAAVAYDMAAIEYRGVNAVTNFDISNYMDKIKKKNDQTQQQQTEAQTET VPNSSDSEEVEVEQQTTTITTPPPSENLHMPPQQHQVQYTPHVSPREEESSSLITIMDHVLEQDLPWSFMYTGLSQFQDP NLAFCKGDDDLVGMFDSAGFEEDIDFLFSTQPGDETESDVNNMSAVLDSVECGDTNGAGGSMMHVDNKQKIVSFASSPSS TTTVSCDYALDL* |
| CDS seq | >KRH13193 ATGAAGAGGTCTCCAGCATCTTCTTGTTCATCATCTACTTCCTCTGTTGGGTTTGAAGCTCCCATTGAAAAAAGAAGGCC TAAGCATCCAAGGAGGAATAATTTGAAGTCACAAAAATGCAAGCAGAACCAAACCACCACTGGTGGCAGAAGAAGCTCTA TCTATAGAGGAGTTACAAGGCATAGGTGGACAGGGAGGTTTGAAGCTCACCTATGGGATAAGAGCTCTTGGAACAACATT CAGAGCAAGAAGGGTCGACAAGTTTATTTGGGGGCATATGATACTGAAGAATCTGCAGCCCGTACCTATGACCTTGCAGC CCTTAAATACTGGGGAAAAGATGCAACCCTGAATTTCCCGATAGAAACTTATACCAAGGAGCTCGAGGAAATGGACAAGG TTTCAAGAGAAGAATATTTGGCTTCTTTGCGGCGCCAAAGCAGTGGCTTTTCTAGAGGCCTGTCTAAGTACCGTGGGGTT GCTAGGCATCATCATAATGGTCGCTGGGAAGCACGAATTGGAAGAGTATGCGGAAACAAGTACCTCTACTTGGGGACATA TAAAACTCAAGAGGAGGCAGCAGTGGCATATGACATGGCAGCAATAGAGTACCGTGGAGTCAATGCAGTGACCAATTTTG ACATAAGCAACTACATGGACAAAATAAAGAAGAAAAATGACCAAACCCAACAACAACAAACAGAAGCACAAACGGAAACA GTTCCTAACTCCTCTGACTCTGAAGAAGTAGAAGTAGAACAACAGACAACAACAATAACCACACCACCCCCATCTGAAAA TCTGCACATGCCACCACAGCAGCACCAAGTTCAATACACCCCCCATGTCTCTCCAAGGGAAGAAGAATCATCATCACTGA TCACAATTATGGACCATGTGCTTGAGCAGGATCTGCCATGGAGCTTCATGTACACTGGCTTGTCTCAGTTTCAAGATCCA AACTTGGCTTTCTGCAAAGGTGATGATGACTTGGTGGGCATGTTTGATAGTGCAGGGTTTGAGGAAGACATTGATTTTCT GTTCAGCACTCAACCTGGTGATGAGACTGAGAGTGATGTCAACAATATGAGCGCAGTTTTGGATAGTGTTGAGTGTGGAG ACACAAATGGGGCTGGTGGAAGCATGATGCATGTGGATAACAAGCAGAAGATAGTATCATTTGCTTCTTCACCATCATCT ACAACTACAGTTTCTTGTGACTATGCTCTAGATCTATGA |