
| Microexon ID | Gm_13:21191140-21191148:+ |
| Species | Glycine max | Coordinates | 13:21191140..21191148 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 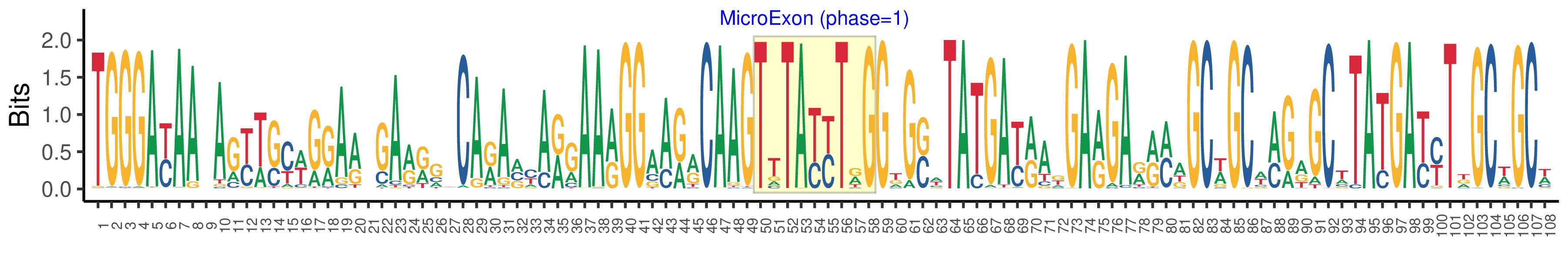 |
| Alignment of exons | 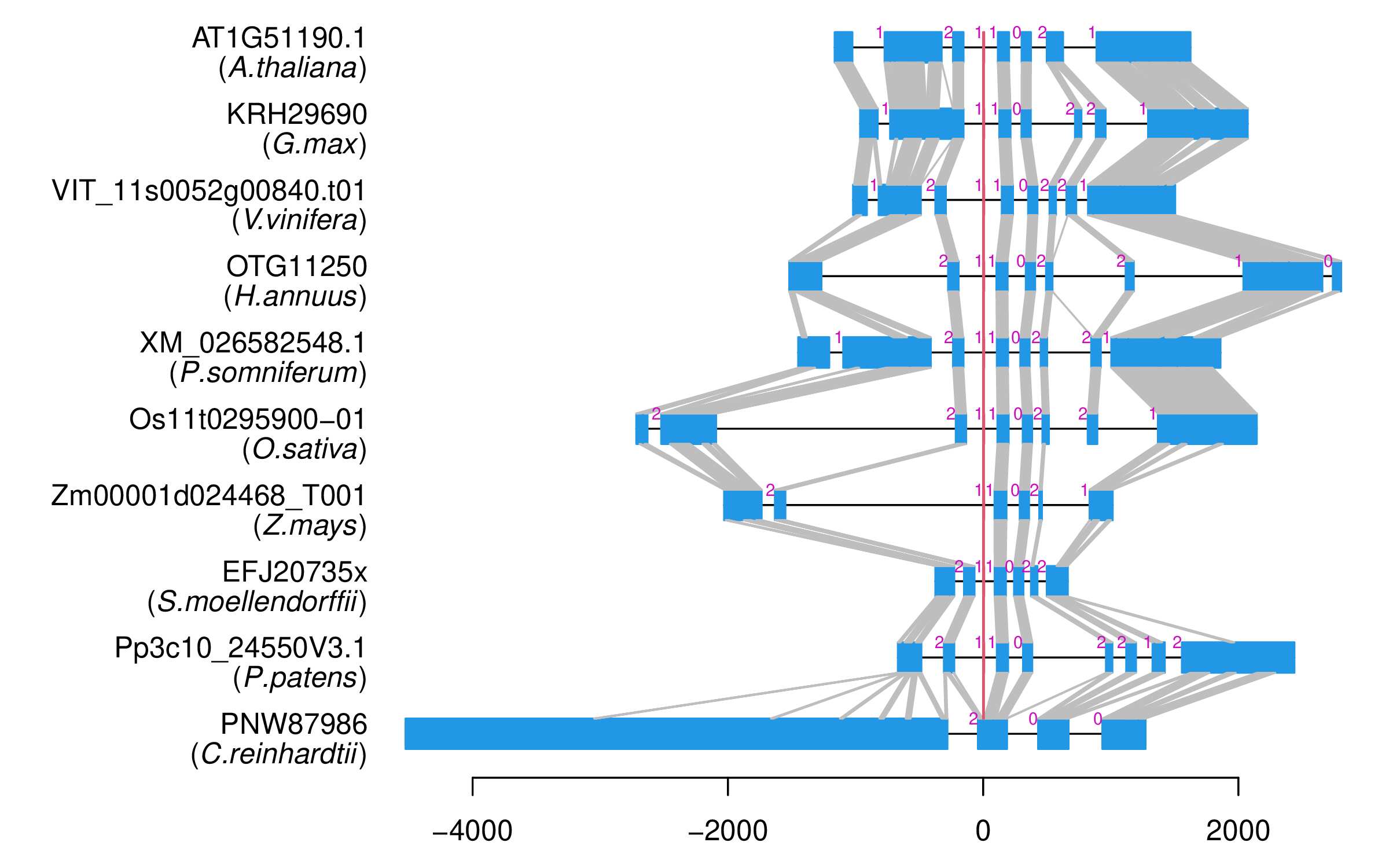 |
| Microexon DNA seq | TTTACCTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAATAGTTGTAGAAGAGAAGGCCAAAGCAGGAAAGGAAGACAAGTTTACCTGGGTGGTTATGACAAGGAAGATAAGGCAGCCAGGGCTTACGATCTCGCAGCT |
| Microexon-tag Amino Acid Seq | WDNSCRREGQSRKGRQVYLGGYDKEDKAARAYDLAA |
| Microexon-tag spanning region | 21190508-21191388 |
| Microexon-tag prediction score | 0.9781 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH19032x |
| Reference Transcript ID | KRH19032 |
| Gene ID | GLYMA_13G096900 |
| Gene Name | NA |
| Transcript ID | KRH19032 |
| Protein ID | KRH19032 |
| Gene ID | GLYMA_13G096900 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 2.6e-12 |
| Motif start | 147 |
| Motif end | 205 |
| Protein seq | >KRH19032 MDSCSSPPNNNSLAFSLSNHFPNPSSSPLSLFHSFTYPSLSLTGSHTADAPPEPIAGGGATNLSIFTGAPKFEDFLGGSS ATATATTCAPPQLPQFSTDNNNHLYDSELKTTIAACFPRAFAAEPTTEPQKPSPKKTVDTFGQRTSIYRGVTRHRWTGRY EAHLWDNSCRREGQSRKGRQVYLGGYDKEDKAARAYDLAALKYWGPTTTTNFPISNYEKELEEMKNMTRQEFVASLRRKS SGFSRGASIYRGVTRHHQHGRWQARIGRVAGNKDLYLGTFSTQEEAAEAYDIAAIKFRGLNAVTNFDMSRYDVKSIANST LPIGGLSGKNKNSTDSASESKSHEASRSDERDPSAASSVTFASQQQPSSSTLSFAIPIKQDPSDYWSILGYHNSPLDNTG IRNTTSVTATSFPSSNNGTTSSLTPFHMEFSNAPTSTGSDNDAAFFSGGGIFVQQQSGHGNGHGSGSSGSSSSSLSCSIP FATPIFSLNSNTSYENSAGYGNWIGPTLHTFQSHAKPSLFQTPIFGME* |
| CDS seq | >KRH19032 ATGGACTCTTGTTCATCACCGCCAAACAACAACTCCCTCGCTTTCTCTCTTTCCAATCACTTTCCCAACCCTTCCTCCTC TCCCCTCTCCCTTTTCCACTCCTTCACCTATCCATCTCTCTCTCTCACAGGAAGCCACACGGCGGATGCACCTCCTGAGC CCATCGCCGGCGGAGGAGCGACCAACCTCTCCATATTCACCGGCGCCCCCAAGTTCGAGGACTTTCTGGGCGGTTCCTCC GCAACAGCCACCGCCACCACGTGTGCACCGCCACAGCTTCCGCAGTTCTCCACCGACAACAACAACCACCTGTACGATTC GGAGCTGAAGACAACAATAGCCGCGTGCTTCCCTCGCGCCTTTGCCGCCGAACCAACCACCGAACCTCAGAAACCCTCTC CAAAGAAAACCGTCGACACCTTCGGCCAACGCACCTCCATCTACCGCGGCGTCACCCGACATAGATGGACGGGAAGATAC GAAGCTCATCTATGGGACAATAGTTGTAGAAGAGAAGGCCAAAGCAGGAAAGGAAGACAAGTTTACCTGGGTGGTTATGA CAAGGAAGATAAGGCAGCCAGGGCTTACGATCTCGCAGCTCTCAAGTACTGGGGTCCAACTACCACCACCAACTTTCCCA TTTCCAACTATGAGAAGGAACTGGAGGAGATGAAGAACATGACCAGGCAAGAGTTTGTTGCTTCTCTACGAAGGAAGAGC AGTGGTTTCTCTAGGGGGGCCTCTATATACAGAGGAGTGACGAGACACCACCAGCATGGCCGATGGCAGGCGAGAATAGG CAGAGTTGCCGGAAACAAAGACCTCTACCTTGGAACTTTCAGCACCCAAGAAGAAGCTGCTGAGGCCTATGACATTGCTG CTATCAAATTCAGGGGATTAAATGCAGTCACAAACTTTGACATGAGTCGCTACGATGTAAAGAGCATTGCAAATAGCACT CTTCCAATTGGAGGTTTATCTGGCAAGAACAAGAACTCCACAGATTCTGCATCTGAGAGCAAGAGCCACGAGGCAAGCCG ATCCGACGAACGAGATCCATCAGCGGCTTCATCCGTGACCTTTGCATCACAGCAACAGCCTTCGAGCTCCACCTTAAGCT TTGCCATACCCATTAAGCAAGACCCTTCAGATTACTGGTCCATCCTGGGGTACCATAATTCTCCCCTTGACAACACTGGC ATCAGGAACACTACTAGTGTTACTGCAACTTCTTTTCCATCCTCCAACAATGGCACTACTAGTAGTTTGACACCCTTCCA CATGGAATTCTCAAATGCCCCCACAAGTACCGGCAGTGATAACGATGCCGCGTTTTTCAGTGGAGGAGGCATCTTTGTTC AGCAACAAAGTGGTCATGGTAATGGTCATGGAAGTGGAAGCAGTGGTTCCTCCTCTTCTTCTTTAAGCTGTTCAATCCCA TTCGCCACGCCCATCTTTTCTCTAAATAGCAATACTAGTTATGAGAACAGTGCTGGTTATGGAAACTGGATTGGACCTAC CCTGCACACATTCCAATCCCATGCAAAACCAAGTCTCTTTCAAACGCCAATATTTGGAATGGAATGA |
| Microexon DNA seq | TTTACCTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGACAATAGTTGTAGAAGAGAAGGCCAAAGCAGGAAAGGAAGACAAGTTTACCTGGGTGGTTATGACAAGGAAGATAAGGCAGCCAGGGCTTACGATCTCGCAGCT |
| Microexon-tag Amino Acid seq | WDNSCRREGQSRKGRQVYLGGYDKEDKAARAYDLAA |
| Transcript ID | KRH19032 |
| Gene ID | Gm.11386 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 2.6e-12 |
| Motif start | 147 |
| Motif end | 205 |
| Protein seq | >KRH19032 MDSCSSPPNNNSLAFSLSNHFPNPSSSPLSLFHSFTYPSLSLTGSHTADAPPEPIAGGGATNLSIFTGAPKFEDFLGGSS ATATATTCAPPQLPQFSTDNNNHLYDSELKTTIAACFPRAFAAEPTTEPQKPSPKKTVDTFGQRTSIYRGVTRHRWTGRY EAHLWDNSCRREGQSRKGRQVYLGGYDKEDKAARAYDLAALKYWGPTTTTNFPISNYEKELEEMKNMTRQEFVASLRRKS SGFSRGASIYRGVTRHHQHGRWQARIGRVAGNKDLYLGTFSTQEEAAEAYDIAAIKFRGLNAVTNFDMSRYDVKSIANST LPIGGLSGKNKNSTDSASESKSHEASRSDERDPSAASSVTFASQQQPSSSTLSFAIPIKQDPSDYWSILGYHNSPLDNTG IRNTTSVTATSFPSSNNGTTSSLTPFHMEFSNAPTSTGSDNDAAFFSGGGIFVQQQSGHGNGHGSGSSGSSSSSLSCSIP FATPIFSLNSNTSYENSAGYGNWIGPTLHTFQSHAKPSLFQTPIFGME* |
| CDS seq | >KRH19032 ATGGACTCTTGTTCATCACCGCCAAACAACAACTCCCTCGCTTTCTCTCTTTCCAATCACTTTCCCAACCCTTCCTCCTC TCCCCTCTCCCTTTTCCACTCCTTCACCTATCCATCTCTCTCTCTCACAGGAAGCCACACGGCGGATGCACCTCCTGAGC CCATCGCCGGCGGAGGAGCGACCAACCTCTCCATATTCACCGGCGCCCCCAAGTTCGAGGACTTTCTGGGCGGTTCCTCC GCAACAGCCACCGCCACCACGTGTGCACCGCCACAGCTTCCGCAGTTCTCCACCGACAACAACAACCACCTGTACGATTC GGAGCTGAAGACAACAATAGCCGCGTGCTTCCCTCGCGCCTTTGCCGCCGAACCAACCACCGAACCTCAGAAACCCTCTC CAAAGAAAACCGTCGACACCTTCGGCCAACGCACCTCCATCTACCGCGGCGTCACCCGACATAGATGGACGGGAAGATAC GAAGCTCATCTATGGGACAATAGTTGTAGAAGAGAAGGCCAAAGCAGGAAAGGAAGACAAGTTTACCTGGGTGGTTATGA CAAGGAAGATAAGGCAGCCAGGGCTTACGATCTCGCAGCTCTCAAGTACTGGGGTCCAACTACCACCACCAACTTTCCCA TTTCCAACTATGAGAAGGAACTGGAGGAGATGAAGAACATGACCAGGCAAGAGTTTGTTGCTTCTCTACGAAGGAAGAGC AGTGGTTTCTCTAGGGGGGCCTCTATATACAGAGGAGTGACGAGACACCACCAGCATGGCCGATGGCAGGCGAGAATAGG CAGAGTTGCCGGAAACAAAGACCTCTACCTTGGAACTTTCAGCACCCAAGAAGAAGCTGCTGAGGCCTATGACATTGCTG CTATCAAATTCAGGGGATTAAATGCAGTCACAAACTTTGACATGAGTCGCTACGATGTAAAGAGCATTGCAAATAGCACT CTTCCAATTGGAGGTTTATCTGGCAAGAACAAGAACTCCACAGATTCTGCATCTGAGAGCAAGAGCCACGAGGCAAGCCG ATCCGACGAACGAGATCCATCAGCGGCTTCATCCGTGACCTTTGCATCACAGCAACAGCCTTCGAGCTCCACCTTAAGCT TTGCCATACCCATTAAGCAAGACCCTTCAGATTACTGGTCCATCCTGGGGTACCATAATTCTCCCCTTGACAACACTGGC ATCAGGAACACTACTAGTGTTACTGCAACTTCTTTTCCATCCTCCAACAATGGCACTACTAGTAGTTTGACACCCTTCCA CATGGAATTCTCAAATGCCCCCACAAGTACCGGCAGTGATAACGATGCCGCGTTTTTCAGTGGAGGAGGCATCTTTGTTC AGCAACAAAGTGGTCATGGTAATGGTCATGGAAGTGGAAGCAGTGGTTCCTCCTCTTCTTCTTTAAGCTGTTCAATCCCA TTCGCCACGCCCATCTTTTCTCTAAATAGCAATACTAGTTATGAGAACAGTGCTGGTTATGGAAACTGGATTGGACCTAC CCTGCACACATTCCAATCCCATGCAAAACCAAGTCTCTTTCAAACGCCAATATTTGGAATGGAATGA |