
| Microexon ID | Gm_12:4101138-4101146:+ |
| Species | Glycine max | Coordinates | 12:4101138..4101146 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 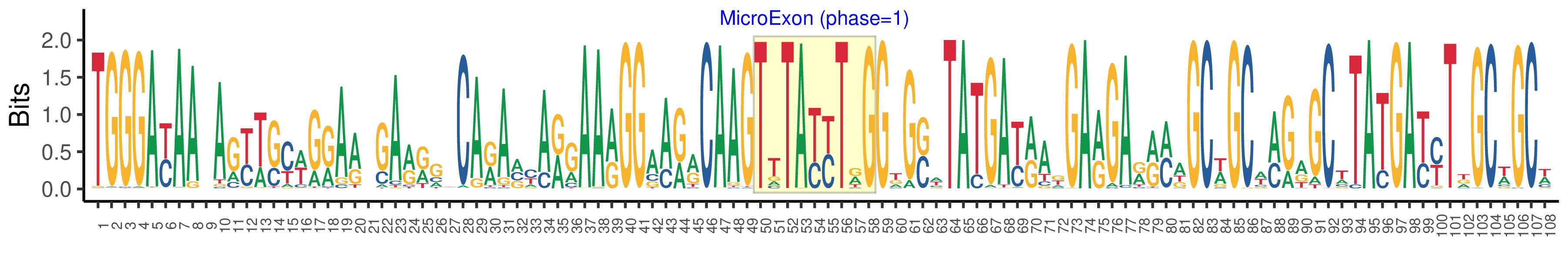 |
| Alignment of exons | 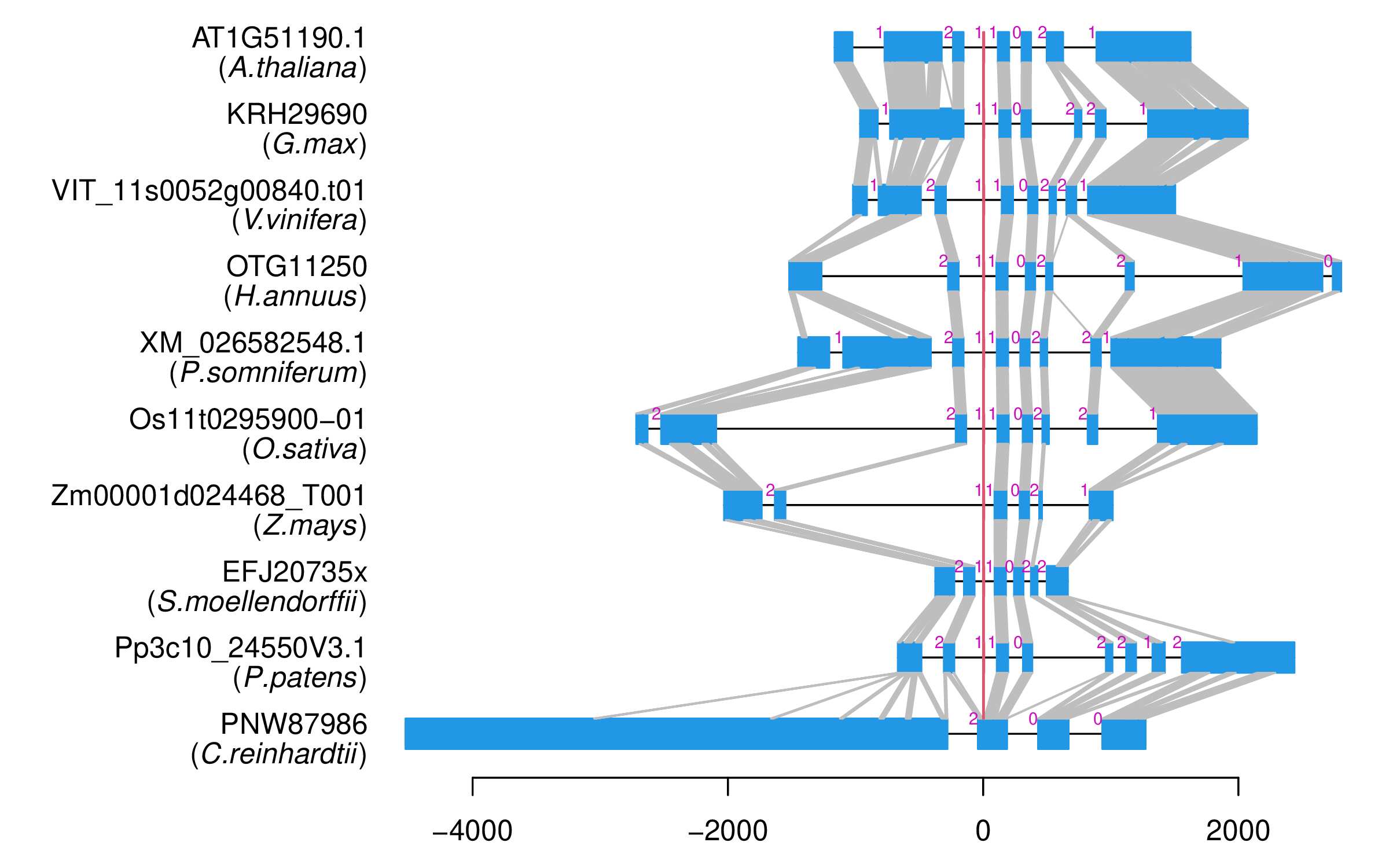 |
| Microexon DNA seq | TTTATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAATAGCTGTAGAAGGGAAGGCCAATCAAGAAAAGGACGCCAAGTTTATTTGGGTGGATATGATAAAGAAGAGAAAGCAGCTAGAGCTTATGATTTAGCTGCA |
| Microexon-tag Amino Acid Seq | WDNSCRREGQSRKGRQVYLGGYDKEEKAARAYDLAA |
| Microexon-tag spanning region | 4100941-4101305 |
| Microexon-tag prediction score | 0.9653 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH24688x |
| Reference Transcript ID | KRH24688 |
| Gene ID | GLYMA_12G056300 |
| Gene Name | NA |
| Transcript ID | KRH24688 |
| Protein ID | KRH24688 |
| Gene ID | GLYMA_12G056300 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 9.8e-13 |
| Motif start | 164 |
| Motif end | 222 |
| Protein seq | >KRH24688 MNNNWLSFPLSPTHSSLPAHDLQATQYHQFSLGLVNENMDNPFQNHDWNLINTHSSNEIPKVADFLGVSKSENQSDLAAL NEIHSNDSDYLFTNNSLVPMQNPVLDTPSNEYQENANSNLQSLTLSMGSGKDSTCETSGENSTNTTVEVAPRRTLDTFGQ RTSIYRGVTRHRWTGRYEAHLWDNSCRREGQSRKGRQVYLGGYDKEEKAARAYDLAALKYWGTSTTTNFPISNYEKELDE MKHMTRQEFVAAIRRKSSGFSRGASMYRGVTRHHQHGRWQARIGRVAGNKDLYLGTFSTEEEAAEAYDIAAIKFRGLNAV TNFDMSRYDVKAILESNTLPIGGGAAKRLKEAQALESSRKREEMIALGSSSTFQYGTSASSSRLHAYPLMQHHHQFEQPQ PLLTLQNHDISSSHFSHQQDPLHHQGYIQTQLQLHQQSGASSYSFQNNAQFYNGYLQNHPALLQGMMNMGSSSSSSSVLE NNNSNNNNNNVGGFVGSGFGMASNATAGNTVGTAEELGLVKVDYDMPAGGYGGWSAADSMQTSNGGVFTMWND* |
| CDS seq | >KRH24688 ATGAACAACAACTGGCTTTCGTTCCCTCTTTCTCCTACTCATTCTTCCTTACCAGCTCATGATCTTCAAGCAACTCAATA TCATCAATTTTCCCTTGGGTTAGTGAACGAGAACATGGATAACCCTTTCCAAAATCATGATTGGAATCTGATTAACACCC ATAGTAGCAACGAAATTCCAAAAGTGGCTGATTTTCTAGGAGTGAGCAAGTCTGAAAATCAGTCAGACCTTGCAGCCTTA AACGAAATTCATTCAAATGATTCAGATTATCTGTTCACAAACAACAGTCTGGTGCCTATGCAAAACCCTGTGTTGGACAC ACCTAGCAATGAGTATCAAGAAAATGCTAATAGTAATTTGCAATCATTGACATTATCCATGGGAAGTGGTAAGGATTCAA CATGTGAAACCAGTGGTGAAAATAGCACAAACACTACTGTTGAAGTTGCACCTAGAAGAACTTTGGATACATTCGGGCAG AGAACATCCATATATCGTGGAGTAACTCGACATAGATGGACTGGAAGGTATGAAGCTCATCTTTGGGATAATAGCTGTAG AAGGGAAGGCCAATCAAGAAAAGGACGCCAAGTTTATTTGGGTGGATATGATAAAGAAGAGAAAGCAGCTAGAGCTTATG ATTTAGCTGCACTGAAGTACTGGGGGACATCCACCACTACCAACTTTCCAATTAGCAACTATGAGAAGGAATTGGATGAA ATGAAACACATGACGAGACAAGAATTTGTTGCCGCCATTAGAAGGAAAAGCAGTGGTTTCTCCAGGGGTGCATCAATGTA TCGTGGAGTTACAAGGCATCACCAACACGGAAGATGGCAAGCAAGGATTGGCAGAGTTGCAGGAAACAAAGATCTTTACT TGGGAACTTTCAGTACTGAGGAAGAGGCTGCAGAAGCATACGACATAGCAGCGATAAAGTTCAGAGGTCTCAACGCTGTC ACAAACTTTGACATGAGCCGCTACGACGTGAAAGCCATTCTTGAAAGCAACACTCTCCCAATAGGAGGAGGCGCTGCAAA GCGTCTGAAAGAAGCTCAAGCTCTAGAATCTTCGAGAAAACGCGAAGAGATGATTGCACTAGGCTCATCTTCCACGTTCC AATACGGAACCTCAGCAAGCTCTTCTAGGCTTCACGCTTACCCTCTAATGCAGCACCACCACCAGTTCGAGCAACCTCAA CCTCTGCTAACTCTTCAAAACCACGACATAAGTTCTTCTCACTTCTCTCACCAGCAAGACCCTTTGCATCATCAGGGTTA CATCCAAACGCAGCTTCAGTTGCACCAGCAGAGTGGCGCTTCTTCTTATAGCTTTCAGAATAATGCTCAGTTCTACAATG GTTACCTTCAGAACCACCCTGCATTGCTTCAGGGAATGATGAACATGGGGTCTTCTTCTTCTTCCTCATCTGTGTTGGAG AATAATAATAGTAACAATAATAATAATAATGTTGGTGGGTTTGTGGGAAGTGGGTTTGGTATGGCTTCGAATGCAACGGC GGGGAACACGGTGGGGACAGCGGAGGAGTTAGGGCTGGTGAAGGTGGACTATGACATGCCGGCTGGAGGTTACGGTGGCT GGTCGGCGGCGGACTCCATGCAGACGTCAAATGGTGGGGTGTTCACAATGTGGAATGATTAA |
| Microexon DNA seq | TTTATTTGG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAATAGCTGTAGAAGGGAAGGCCAATCAAGAAAAGGACGCCAAGTTTATTTGGGTGGATATGATAAAGAAGAGAAAGCAGCTAGAGCTTATGATTTAGCTGCA |
| Microexon-tag Amino Acid seq | WDNSCRREGQSRKGRQVYLGGYDKEEKAARAYDLAA |
| Transcript ID | Gm.8404.1 |
| Gene ID | Gm.8404 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 9.8e-13 |
| Motif start | 164 |
| Motif end | 222 |
| Protein seq | >Gm.8404.1 MNNNWLSFPLSPTHSSLPAHDLQATQYHQFSLGLVNENMDNPFQNHDWNLINTHSSNEIPKVADFLGVSKSENQSDLAAL NEIHSNDSDYLFTNNSLVPMQNPVLDTPSNEYQENANSNLQSLTLSMGSGKDSTCETSGENSTNTTVEVAPRRTLDTFGQ RTSIYRGVTRHRWTGRYEAHLWDNSCRREGQSRKGRQVYLGGYDKEEKAARAYDLAALKYWGTSTTTNFPISNYEKELDE MKHMTRQEFVAAIRRKSSGFSRGASMYRGVTRHHQHGRWQARIGRVAGNKDLYLGTFSTEEEAAEAYDIAAIKFRGLNAV TNFDMSRYDVKAILESNTLPIGGGAAKRLKEAQALESSRKREEMIALGSSSTFQYGTSASSSRLHAYPLMQHHHQFEQPQ PLLTLQNHDISSSHFSHQQDPLHHQGYIQTQLQLHQQSGASSYSFQNNAQFYNGYLQNHPALLQGMMNMGSSSSSSSVLE NNNSNNNNNNVGGFVGSGFGMASNATAGNTVGTAEELGLVKVDYDMPAGGYGGWSAADSMQTSNGGVFTMWND* |
| CDS seq | >Gm.8404.1 ATGAACAACAACTGGCTTTCGTTCCCTCTTTCTCCTACTCATTCTTCCTTACCAGCTCATGATCTTCAAGCAACTCAATA TCATCAATTTTCCCTTGGGTTAGTGAACGAGAACATGGATAACCCTTTCCAAAATCATGATTGGAATCTGATTAACACCC ATAGTAGCAACGAAATTCCAAAAGTGGCTGATTTTCTAGGAGTGAGCAAGTCTGAAAATCAGTCAGACCTTGCAGCCTTA AACGAAATTCATTCAAATGATTCAGATTATCTGTTCACAAACAACAGTCTGGTGCCTATGCAAAACCCTGTGTTGGACAC ACCTAGCAATGAGTATCAAGAAAATGCTAATAGTAATTTGCAATCATTGACATTATCCATGGGAAGTGGTAAGGATTCAA CATGTGAAACCAGTGGTGAAAATAGCACAAACACTACTGTTGAAGTTGCACCTAGAAGAACTTTGGATACATTCGGGCAG AGAACATCCATATATCGTGGAGTAACTCGACATAGATGGACTGGAAGGTATGAAGCTCATCTTTGGGATAATAGCTGTAG AAGGGAAGGCCAATCAAGAAAAGGACGCCAAGTTTATTTGGGTGGATATGATAAAGAAGAGAAAGCAGCTAGAGCTTATG ATTTAGCTGCACTGAAGTACTGGGGGACATCCACCACTACCAACTTTCCAATTAGCAACTATGAGAAGGAATTGGATGAA ATGAAACACATGACGAGACAAGAATTTGTTGCCGCCATTAGAAGGAAAAGCAGTGGTTTCTCCAGGGGTGCATCAATGTA TCGTGGAGTTACAAGGCATCACCAACACGGAAGATGGCAAGCAAGGATTGGCAGAGTTGCAGGAAACAAAGATCTTTACT TGGGAACTTTCAGTACTGAGGAAGAGGCTGCAGAAGCATACGACATAGCAGCGATAAAGTTCAGAGGTCTCAACGCTGTC ACAAACTTTGACATGAGCCGCTACGACGTGAAAGCCATTCTTGAAAGCAACACTCTCCCAATAGGAGGAGGCGCTGCAAA GCGTCTGAAAGAAGCTCAAGCTCTAGAATCTTCGAGAAAACGCGAAGAGATGATTGCACTAGGCTCATCTTCCACGTTCC AATACGGAACCTCAGCAAGCTCTTCTAGGCTTCACGCTTACCCTCTAATGCAGCACCACCACCAGTTCGAGCAACCTCAA CCTCTGCTAACTCTTCAAAACCACGACATAAGTTCTTCTCACTTCTCTCACCAGCAAGACCCTTTGCATCATCAGGGTTA CATCCAAACGCAGCTTCAGTTGCACCAGCAGAGTGGCGCTTCTTCTTATAGCTTTCAGAATAATGCTCAGTTCTACAATG GTTACCTTCAGAACCACCCTGCATTGCTTCAGGGAATGATGAACATGGGGTCTTCTTCTTCTTCCTCATCTGTGTTGGAG AATAATAATAGTAACAATAATAATAATAATGTTGGTGGGTTTGTGGGAAGTGGGTTTGGTATGGCTTCGAATGCAACGGC GGGGAACACGGTGGGGACAGCGGAGGAGTTAGGGCTGGTGAAGGTGGACTATGACATGCCGGCTGGAGGTTACGGTGGCT GGTCGGCGGCGGACTCCATGCAGACGTCAAATGGTGGGGTGTTCACAATGTGGAATGATTAA |