
| Microexon ID | At_1:29993080-29993088:- |
| Species | Arabidopsis thaliana | Coordinates | 1:29993080..29993088 |
| Microexon Cluster ID | MEP21 |
| Size | 9 |
| Phase | 1 |
| Pfam Domain Motif | AP2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,9,50 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TGGGAYAAYAGYWSYWGRARWGARRGYCARAVYARGAARGGAARRCAAGTTTAYYTRGGKGSWTATGAYRAKGARGARRMWGCWGCWMGRGCWTATGAYYTDGCWGCW |
| Logo of Microexon-tag DNA Seq | 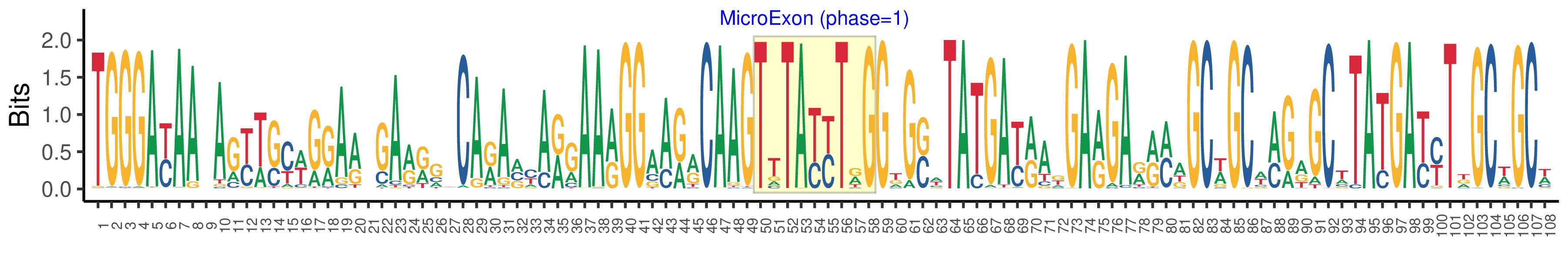 |
| Alignment of exons | 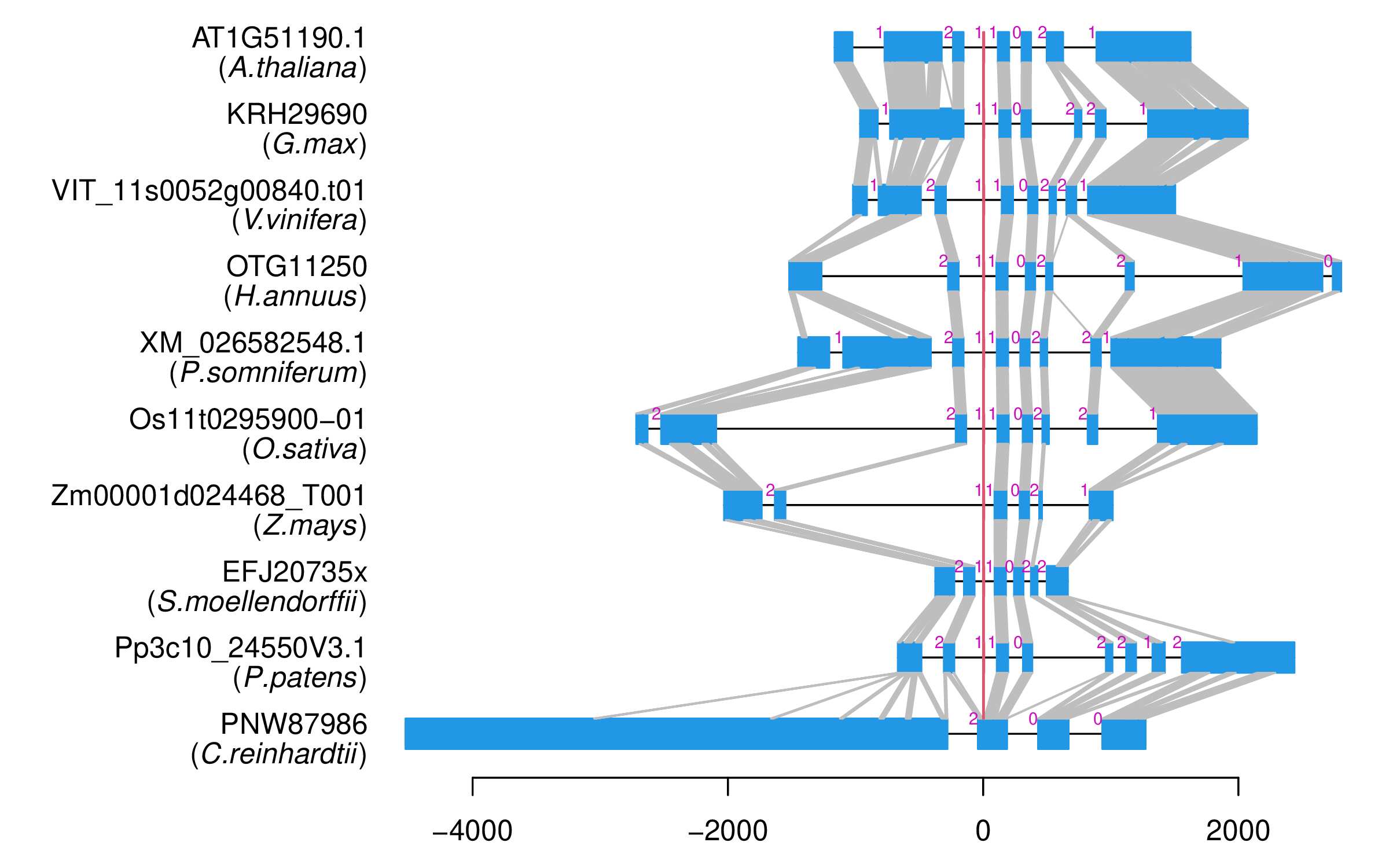 |
| Microexon DNA seq | TTTATCTAG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAAGAACAGCTGGAACGATACACAGACCAAGAAAGGACGTCAAGTTTATCTAGGGGCTTACGACGAAGAAGAAGCAGCAGCACGTGCCTACGACTTAGCAGCA |
| Microexon-tag Amino Acid Seq | WDKNSWNDTQTKKGRQVYLGAYDEEEAAARAYDLAA |
| Microexon-tag spanning region | 29992677-29993370 |
| Microexon-tag prediction score | 0.9628 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | AT1G79700.1x |
| Reference Transcript ID | AT1G79700.1 |
| Gene ID | AT1G79700 |
| Gene Name | NA |
| Transcript ID | AT1G79700.1 |
| Protein ID | AT1G79700.1 |
| Gene ID | AT1G79700 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 1.3e-13 |
| Motif start | 53 |
| Motif end | 110 |
| Protein seq | >AT1G79700.1 MAKVSGRSKKTIVDDEISDKTASASESASIALTSKRKRKSPPRNAPLQRSSPYRGVTRHRWTGRYEAHLWDKNSWNDTQT KKGRQVYLGAYDEEEAAARAYDLAALKYWGRDTLLNFPLPSYDEDVKEMEGQSKEEYIGSLRRKSSGFSRGVSKYRGVAR HHHNGRWEARIGRVFATQEEAAIAYDIAAIEYRGLNAVTNFDVSRYLNPNAAADKADSDSKPIRSPSREPESSDDNKSPK SEEVIEPSTSPEVIPTRRSFPDDIQTYFGCQDSGKLATEEDVIFDCFNSYINPGFYNEFDYGP* |
| CDS seq | >AT1G79700.1 ATGGCAAAAGTCTCTGGGAGGAGCAAGAAAACAATCGTTGACGATGAAATCAGCGATAAAACAGCGTCTGCGTCTGAGTC TGCGTCCATTGCCTTAACATCCAAACGCAAACGTAAGTCGCCGCCTCGAAACGCTCCTCTTCAACGCAGCTCCCCTTACA GAGGCGTCACAAGGCATAGATGGACTGGGAGATACGAAGCGCATTTGTGGGATAAGAACAGCTGGAACGATACACAGACC AAGAAAGGACGTCAAGTTTATCTAGGGGCTTACGACGAAGAAGAAGCAGCAGCACGTGCCTACGACTTAGCAGCATTGAA GTACTGGGGACGAGACACACTCTTGAACTTCCCTTTGCCGAGTTATGACGAAGACGTCAAAGAAATGGAAGGCCAATCCA AGGAAGAGTATATTGGATCATTGAGAAGAAAAAGTAGTGGATTTTCTCGCGGTGTATCAAAATACAGAGGCGTTGCAAGG CATCACCATAATGGGAGATGGGAAGCTAGAATTGGAAGGGTGTTTGCCACGCAAGAAGAAGCAGCAATCGCCTACGACAT CGCGGCAATAGAGTACCGTGGACTTAACGCCGTTACCAATTTCGACGTCAGCCGTTATCTAAACCCTAACGCCGCCGCGG ATAAAGCCGATTCCGATTCTAAGCCCATTCGAAGCCCTAGTCGCGAGCCCGAATCGTCGGATGATAACAAATCTCCGAAA TCAGAGGAAGTAATCGAACCATCTACATCGCCGGAAGTGATTCCAACTCGCCGGAGCTTCCCCGACGATATCCAGACGTA TTTTGGGTGTCAAGATTCCGGCAAGTTAGCGACTGAGGAAGACGTAATATTCGATTGTTTCAATTCTTATATAAATCCTG GCTTCTATAACGAGTTTGATTATGGACCTTAA |
| Microexon DNA seq | TTTATCTAG |
| Microexon Amino Acid seq | VYLG |
| Microexon-tag DNA Seq | TGGGATAAGAACAGCTGGAACGATACACAGACCAAGAAAGGACGTCAAGTTTATCTAGGGGCTTACGACGAAGAAGAAGCAGCAGCACGTGCCTACGACTTAGCAGCA |
| Microexon-tag Amino Acid seq | WDKNSWNDTQTKKGRQVYLGAYDEEEAAARAYDLAA |
| Transcript ID | AT1G79700.1 |
| Gene ID | At.7270 |
| Gene Name | NA |
| Pfam domain motif | AP2 |
| Motif E-value | 1.3e-13 |
| Motif start | 53 |
| Motif end | 110 |
| Protein seq | >AT1G79700.1 MAKVSGRSKKTIVDDEISDKTASASESASIALTSKRKRKSPPRNAPLQRSSPYRGVTRHRWTGRYEAHLWDKNSWNDTQT KKGRQVYLGAYDEEEAAARAYDLAALKYWGRDTLLNFPLPSYDEDVKEMEGQSKEEYIGSLRRKSSGFSRGVSKYRGVAR HHHNGRWEARIGRVFATQEEAAIAYDIAAIEYRGLNAVTNFDVSRYLNPNAAADKADSDSKPIRSPSREPESSDDNKSPK SEEVIEPSTSPEVIPTRRSFPDDIQTYFGCQDSGKLATEEDVIFDCFNSYINPGFYNEFDYGP* |
| CDS seq | >AT1G79700.1 ATGGCAAAAGTCTCTGGGAGGAGCAAGAAAACAATCGTTGACGATGAAATCAGCGATAAAACAGCGTCTGCGTCTGAGTC TGCGTCCATTGCCTTAACATCCAAACGCAAACGTAAGTCGCCGCCTCGAAACGCTCCTCTTCAACGCAGCTCCCCTTACA GAGGCGTCACAAGGCATAGATGGACTGGGAGATACGAAGCGCATTTGTGGGATAAGAACAGCTGGAACGATACACAGACC AAGAAAGGACGTCAAGTTTATCTAGGGGCTTACGACGAAGAAGAAGCAGCAGCACGTGCCTACGACTTAGCAGCATTGAA GTACTGGGGACGAGACACACTCTTGAACTTCCCTTTGCCGAGTTATGACGAAGACGTCAAAGAAATGGAAGGCCAATCCA AGGAAGAGTATATTGGATCATTGAGAAGAAAAAGTAGTGGATTTTCTCGCGGTGTATCAAAATACAGAGGCGTTGCAAGG CATCACCATAATGGGAGATGGGAAGCTAGAATTGGAAGGGTGTTTGCCACGCAAGAAGAAGCAGCAATCGCCTACGACAT CGCGGCAATAGAGTACCGTGGACTTAACGCCGTTACCAATTTCGACGTCAGCCGTTATCTAAACCCTAACGCCGCCGCGG ATAAAGCCGATTCCGATTCTAAGCCCATTCGAAGCCCTAGTCGCGAGCCCGAATCGTCGGATGATAACAAATCTCCGAAA TCAGAGGAAGTAATCGAACCATCTACATCGCCGGAAGTGATTCCAACTCGCCGGAGCTTCCCCGACGATATCCAGACGTA TTTTGGGTGTCAAGATTCCGGCAAGTTAGCGACTGAGGAAGACGTAATATTCGATTGTTTCAATTCTTATATAAATCCTG GCTTCTATAACGAGTTTGATTATGGACCTTAA |