
| Microexon ID | Bn_scaffoldC03:67503161-67503168:+ |
| Species | Brassica Napus | Coordinates | scaffoldC03:67503161..67503168 |
| Microexon Cluster ID | MEP18 |
| Size | 8 |
| Phase | 1 |
| Pfam Domain Motif | DEAD |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,8,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | CCTGCWCTRGCAAARMCAGGMATTGTKCTTGTTGTTTSTCCYTTRATAGCHYTRATGGARAAYCARGTTAYGRSMTTGAARGARAAAGGRRTTSCWGCTGAATWTCTC |
| Logo of Microexon-tag DNA Seq | 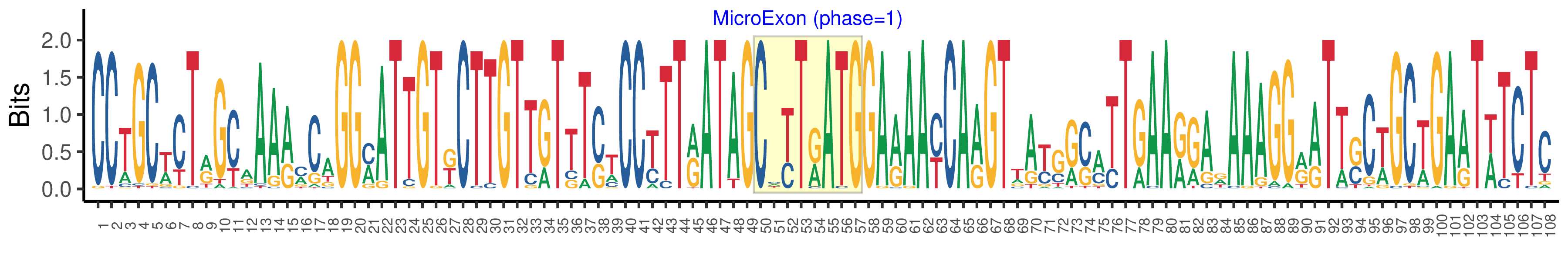 |
| Alignment of exons | 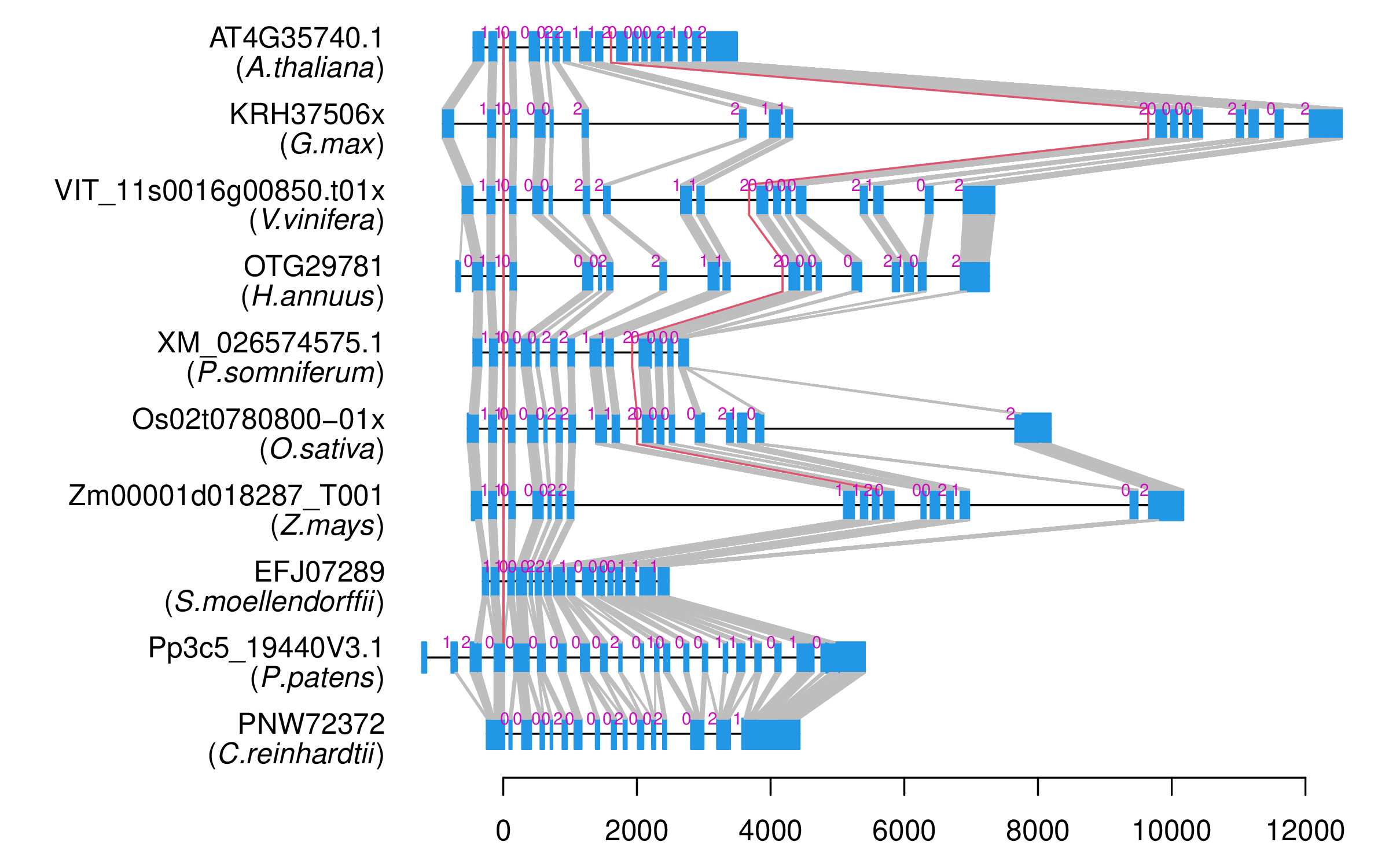 |
| Microexon DNA seq | CTTTGATG |
| Microexon Amino Acid seq | ALM |
| Microexon-tag DNA Seq | CCTGCGTTGGCTAAACCTGGAATCGTGCTTGTTGTTTCTCCTTTGATTGCTTTGATGGAGAATCAAGTGATGGCGTTGAAGGAGAAAGGTATTGCTGCTGAGTATCTC |
| Microexon-tag Amino Acid Seq | PALAKPGIVLVVSPLIALMENQVMALKEKGIAAEYL |
| Microexon-tag spanning region | 67503012-67503310 |
| Microexon-tag prediction score | 0.9664 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | BnaC03T0699800ZSx |
| Reference Transcript ID | BnaC03T0699800ZS |
| Gene ID | BnaC03G0699800ZS |
| Gene Name | NA |
| Transcript ID | BnaC03T0699800ZS |
| Protein ID | |
| Gene ID | BnaC03G0699800ZS |
| Gene Name | |
| Pfam domain motif | DEAD |
| Motif E-value | 2.00E-25 |
| Motif start | 41 |
| Motif end | 205 |
| Protein seq | >BnaC03T0699800ZS MKKSPLPVQNVNRSDKNVSGKEVLVKLLRWHFGHADFRGKQLEAIQAVVSGRDCFCLMPTGGGKSICYQIPALAKPGIVL VVSPLIALMENQVMALKEKGIAAEYLSSTQATHVRNKIHEDLDSGKPSVRLLYVTPELIATKGFMLKLRKLHDRGLLNLI AIDEAHCISSWGHDFRPSYRQLSTLRDSLADVPVLALTATAAPKVQTDVIESLSLRNPLVLKSSFNRPNIFYEVRYKDLI DNAYTDLCNLLKSCGNICAIIYCLERTTCDDLSLHLSSTGISSAAYHAGLNSNLRSTVLDDWLSSKKQVIVATVAFGMGI DKKDVRMVCHFNVPKSMESFYQESGRAGRDQLPSRSVLYYGVDDRKKMEFLLRNSENKKSQSSSSKKPTSDFEQIVRYCE GSGCRRKKILESFGEEFPVQQCNKTCDACKHPNQVARSLEELTTTASRRHNSSRVFITSSSDNKTNDGQYSEFWNRNEDG SNSDEEISDSDDGADVVKSLAGPKLSRKLGVDEKLVLLERAEEKYNESNKQVKKSEKNAISETLRESSKQRLLNELTKVL QLLGVKEIESQNASEFLENECYRKYSKAGKSFYYSQIASTVRWLGTATRDELMTRLSSMAREEEPSGEPILVTEPVENIR RQKHIYSRATGR* |
| CDS seq | >BnaC03T0699800ZS ATGAAGAAATCGCCGTTGCCGGTGCAAAACGTTAACCGCTCCGATAAGAACGTTTCAGGAAAAGAAGTATTGGTGAAGCT ATTGCGATGGCACTTCGGCCATGCAGATTTCAGAGGGAAGCAGCTAGAGGCGATTCAAGCGGTTGTGTCAGGAAGGGATT GCTTTTGCTTGATGCCGACGGGAGGAGGGAAATCGATTTGTTATCAGATACCTGCGTTGGCTAAACCTGGAATCGTGCTT GTTGTTTCTCCTTTGATTGCTTTGATGGAGAATCAAGTGATGGCGTTGAAGGAGAAAGGTATTGCTGCTGAGTATCTCTC GTCCACTCAAGCTACACATGTTAGGAACAAGATCCACGAGGACCTTGATTCTGGAAAACCTTCTGTGAGATTGCTGTATG TAACTCCAGAATTGATCGCGACAAAAGGATTTATGCTGAAGCTGAGAAAGCTCCATGACAGGGGTTTACTGAATCTTATA GCCATAGACGAGGCACATTGCATCTCATCATGGGGCCACGACTTCAGACCTAGCTATCGTCAACTTTCAACATTAAGAGA TTCTTTGGCTGATGTTCCAGTCTTGGCTTTAACCGCTACAGCTGCTCCTAAGGTACAAACAGATGTTATTGAGTCCTTGA GTCTGCGGAATCCTTTGGTCCTCAAGTCTTCTTTCAATCGCCCAAATATCTTCTATGAAGTTCGGTACAAAGATCTTATA GATAATGCTTATACTGATTTGTGCAACTTGCTCAAGTCATGTGGAAATATATGTGCAATCATCTATTGCCTTGAGCGTAC AACCTGTGATGACTTGTCTCTTCATCTTTCCAGTACTGGGATCTCTTCTGCTGCCTATCATGCAGGACTGAATAGTAATC TGAGAAGTACCGTATTAGATGACTGGCTGTCCTCGAAGAAGCAAGTTATTGTCGCCACCGTGGCTTTCGGAATGGGTATA GATAAGAAGGATGTCAGGATGGTTTGCCACTTCAACGTTCCAAAATCGATGGAATCTTTCTACCAAGAGTCCGGCAGAGC AGGTCGGGATCAACTACCTTCAAGAAGTGTATTATACTATGGCGTAGACGATAGAAAGAAAATGGAGTTTCTACTACGAA ACTCGGAGAATAAGAAATCCCAATCCTCATCCTCAAAGAAGCCTACATCCGACTTCGAGCAGATTGTGAGATACTGCGAA GGTTCTGGATGCCGAAGGAAAAAGATTCTTGAGAGCTTTGGTGAAGAGTTTCCCGTACAGCAATGCAACAAAACGTGTGA TGCATGTAAGCATCCAAATCAAGTTGCTCGTAGCTTGGAGGAGCTCACGACTACCGCCTCCCGAAGACACAACTCTTCTC GAGTTTTCATCACCAGCTCGAGTGATAATAAGACCAATGACGGGCAGTACTCTGAGTTCTGGAATCGTAATGAAGATGGA AGCAATTCCGACGAGGAAATATCAGATTCAGATGATGGAGCTGATGTTGTCAAAAGCCTAGCAGGACCTAAACTATCGAG GAAGCTGGGAGTAGATGAGAAACTGGTTTTACTGGAGCGGGCTGAGGAAAAGTACAACGAAAGCAATAAACAGGTCAAAA AGTCAGAGAAGAATGCAATATCTGAAACACTAAGGGAATCAAGCAAGCAGAGACTCTTGAATGAGCTTACAAAAGTACTT CAACTGCTCGGTGTAAAAGAGATTGAGTCCCAAAACGCATCTGAGTTTCTTGAAAACGAGTGCTATAGAAAATACAGCAA AGCAGGGAAATCGTTTTACTACTCACAGATAGCGAGTACTGTGAGATGGTTGGGAACCGCAACCAGAGACGAGCTAATGA CCCGACTTAGCTCAATGGCCAGAGAAGAAGAGCCATCAGGAGAACCAATCCTGGTGACTGAACCAGTCGAGAACATAAGA AGACAGAAACACATATACAGCCGAGCTACAGGTCGATGA |
Bn_scaffoldC03:67503161-67503168:+ does not have available information here.