
| Microexon ID | Sl_6:120141-120148:+ |
| Species | Solanum Lycopersicum | Coordinates | 6:120141..120148 |
| Microexon Cluster ID | MEP17 |
| Size | 8 |
| Phase | 1 |
| Pfam Domain Motif | IP_trans |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,8,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | CTYRYSRTGCAAGAGGAAKCYTGGWATGCRTAYCCAARRWGCARAACAGTRATCAAGTGCCCRYAYTTYASCAAGTKYWCWYTRACCATTGAMACYRTYMAYARRSCY |
| Logo of Microexon-tag DNA Seq | 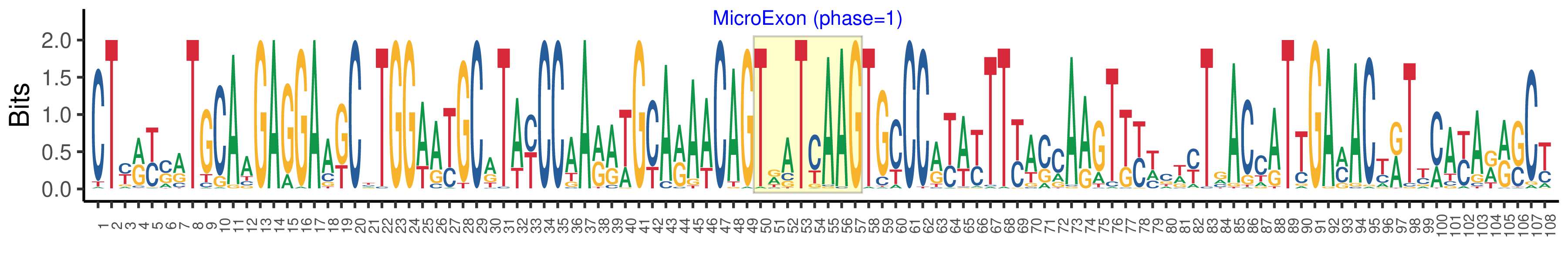 |
| Alignment of exons | 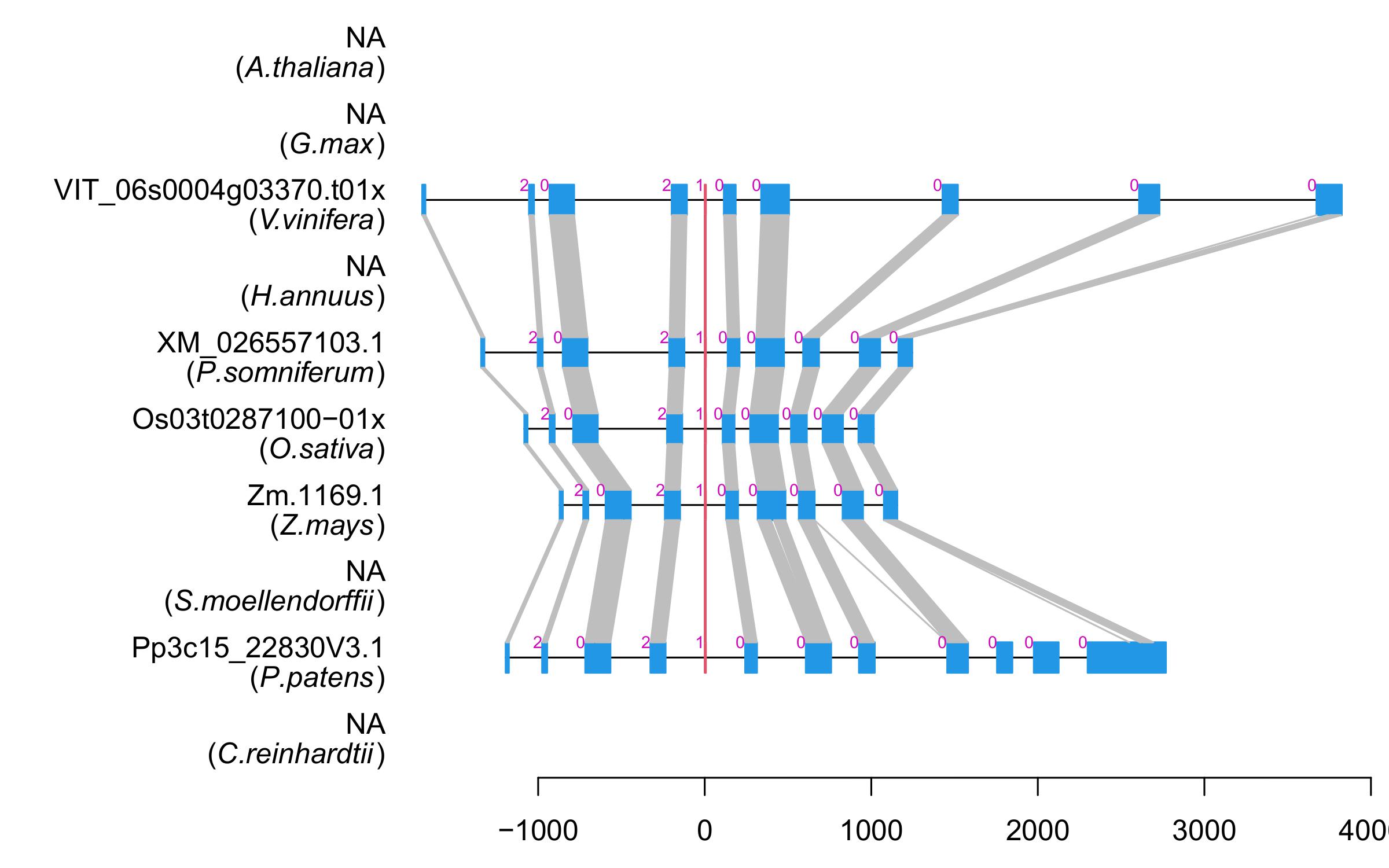 |
| Microexon DNA seq | TTGTTAAG |
| Microexon Amino Acid seq | VVK |
| Microexon-tag DNA Seq | CTTGTGATGCAAGAGGAAGCCTGGAATGCATATCCTAAATGTCGATCAGTTGTTAAGTTACCATATTTTTCGAAGTTCTTTATGAGTGTCGACACTATCCACAAAGCT |
| Microexon-tag Amino Acid Seq | LVMQEEAWNAYPKCRSVVKLPYFSKFFMSVDTIHKA |
| Microexon-tag spanning region | 119972-120295 |
| Microexon-tag prediction score | 0.9321 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Solyc06g005100.3.1x |
| Reference Transcript ID | Solyc06g005100.3.1 |
| Gene ID | Solyc06g005100.3 |
| Gene Name | NA |
| Transcript ID | Solyc06g005100.3.1 |
| Protein ID | |
| Gene ID | Solyc06g005100.3 |
| Gene Name | |
| Pfam domain motif | IP_trans |
| Motif E-value | 1.80E-51 |
| Motif start | 1 |
| Motif end | 163 |
| Protein seq | >Solyc06g005100.3.1 MQEEAWNAYPKCRSVVKLPYFSKFFMSVDTIHKADNGYSENVHGLSENQLAVRQVEFIDIASAATDYWSYVVGRNNVDFS NFQSARCGRGPLLEGWQDLCNPVMTTYKLVTVDAPYWGFGSRLEQALMAGERSLFLESHRNCFAWIDEWFGLTVEVLREL ERQSDYALNMKLGRPCSAESWMTQEESPLEGEKSVA* |
| CDS seq | >Solyc06g005100.3.1 ATGCAAGAGGAAGCCTGGAATGCATATCCTAAATGTCGATCAGTTGTTAAGTTACCATATTTTTCGAAGTTCTTTATGAG TGTCGACACTATCCACAAAGCTGACAATGGGTATTCTGAGAATGTGCATGGCCTGAGTGAGAACCAACTGGCAGTTAGGC AGGTGGAATTTATTGATATTGCTTCAGCTGCCACTGATTATTGGAGTTACGTAGTTGGAAGAAACAATGTTGATTTCTCT AATTTCCAATCAGCAAGATGTGGTCGTGGACCACTCTTGGAAGGCTGGCAGGACCTCTGTAATCCCGTTATGACAACATA CAAACTGGTGACCGTAGATGCTCCATACTGGGGGTTTGGGAGCAGATTGGAACAAGCTTTGATGGCGGGGGAAAGGTCAC TTTTCCTAGAAAGCCATCGTAATTGCTTTGCCTGGATTGATGAATGGTTTGGGTTGACAGTAGAGGTGTTGCGTGAGTTG GAGCGACAGAGTGATTATGCATTGAATATGAAACTTGGCCGGCCTTGCTCGGCAGAGAGTTGGATGACACAAGAGGAATC TCCACTTGAAGGCGAAAAGTCTGTTGCATGA |
Sl_6:120141-120148:+ does not have available information here.