
| Microexon ID | Ps_NC_039366.1:28283026-28283033:+ |
| Species | Papaver somniferum | Coordinates | NC_039366.1:28283026..28283033 |
| Microexon Cluster ID | MEP17 |
| Size | 8 |
| Phase | 1 |
| Pfam Domain Motif | IP_trans |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,8,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | CTYRYSRTGCAAGAGGAAKCYTGGWATGCRTAYCCAARRWGCARAACAGTRATCAAGTGCCCRYAYTTYASCAAGTKYWCWYTRACCATTGAMACYRTYMAYARRSCY |
| Logo of Microexon-tag DNA Seq | 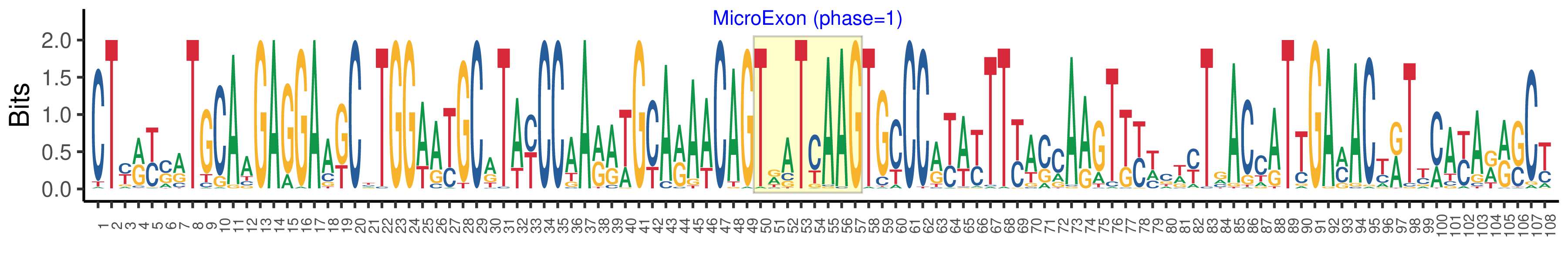 |
| Alignment of exons | 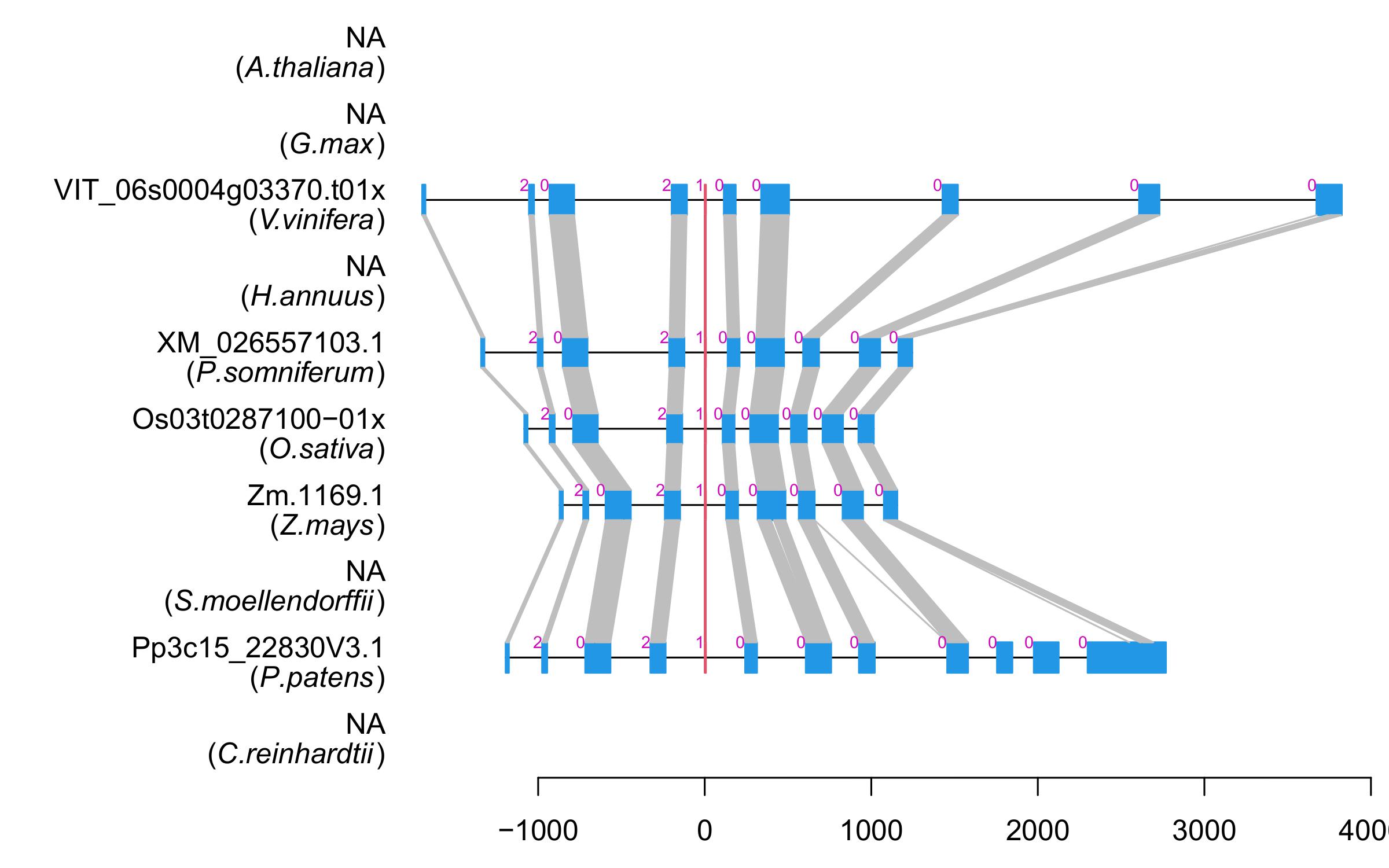 |
| Microexon DNA seq | TGATTAAG |
| Microexon Amino Acid seq | VIK |
| Microexon-tag DNA Seq | CTCCTCATGCAGGAAGAAGCTTGGAATGCTTATCCAAGATGTAAAACAGTGATTAAGTGCCCGTACTTCGAGAGATTCAACTTATCCATTGAAACTATTCATAAGGCT |
| Microexon-tag Amino Acid Seq | LLMQEEAWNAYPRCKTVIKCPYFERFNLSIETIHKA |
| Microexon-tag spanning region | 28282856-28283214 |
| Microexon-tag prediction score | 0.9363 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026557103.1x |
| Reference Transcript ID | XM_026557103.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026557103.1 |
| Protein ID | XP_026412888.1 |
| Gene ID | LOC113308637 |
| Gene Name | NA |
| Pfam domain motif | IP_trans |
| Motif E-value | 6.3e-103 |
| Motif start | 3 |
| Motif end | 247 |
| Protein seq | >XP_026412888.1 MVQLREFRIVMPMSLDEYQVAQMYMVMKMQQQSTSNNEGVEVLENRPFKDDVFGKGQYTSKVYHLQSKIPTWLTTFAPAD ALLMQEEAWNAYPRCKTVIKCPYFERFNLSIETIHKADNGNSENVHGLSKEQLATREVVIIDIASTVTDYWSYAIGTSPV DLSKFKSSRTNRGPLLEGWQDLCEPVMTAYKLVTVDAPYWGFGYRLEQALLSAERALFLESHRNCFAWIDDWYGMTMEQI RELEKQSDSSLNEKLGKVASVDDTQGLDLNDSDKNMKQLS* |
| CDS seq | >XM_026557103.1 ATGGTTCAACTCAGAGAATTTCGGATTGTCATGCCTATGTCGCTTGATGAGTATCAGGTAGCCCAGATGTATATGGTAAT GAAGATGCAACAGCAAAGTACGTCGAATAATGAAGGTGTGGAGGTGTTAGAAAACAGACCATTTAAAGATGATGTGTTTG GGAAGGGTCAGTACACATCTAAAGTGTATCACTTGCAGAGCAAAATTCCTACTTGGCTCACTACTTTTGCACCCGCAGAT GCTCTCCTCATGCAGGAAGAAGCTTGGAATGCTTATCCAAGATGTAAAACAGTGATTAAGTGCCCGTACTTCGAGAGATT CAACTTATCCATTGAAACTATTCATAAGGCTGACAATGGGAACTCAGAAAATGTGCATGGTTTAAGCAAAGAACAGCTAG CGACCAGGGAAGTAGTGATTATTGATATAGCTTCAACTGTGACGGATTATTGGAGCTATGCTATTGGTACCAGCCCTGTG GATTTGAGCAAATTCAAATCATCGAGGACAAACCGCGGTCCGCTATTGGAGGGATGGCAGGATCTGTGTGAACCAGTAAT GACAGCATATAAACTGGTGACGGTAGATGCTCCATACTGGGGTTTTGGGTATCGACTGGAACAAGCTTTACTTTCCGCTG AAAGGGCACTCTTTCTGGAAAGCCATAGGAACTGCTTTGCTTGGATTGATGATTGGTATGGGATGACAATGGAGCAAATA CGAGAACTTGAAAAACAAAGTGATTCGTCACTGAATGAGAAACTGGGGAAGGTAGCTTCAGTGGACGACACACAAGGTCT CGACTTAAATGACAGTGACAAGAATATGAAGCAACTAAGCTAG |
| Microexon DNA seq | TGATTAAG |
| Microexon Amino Acid seq | VIK |
| Microexon-tag DNA Seq | ATGTGTTTGGGAAGGGTCAGTACACATCTAAAGTGTATCACTTGCAGAGTGATTAAGTGCCCGTACTTCGAGAGATTCAACTTATCCATTGAAACTATTCATAAGGCT |
| Microexon-tag Amino Acid seq | MCLGRVSTHLKCITCRVIKCPYFERFNLSIETIHKA |
| Transcript ID | Ps.54159.2 |
| Gene ID | Ps.54159 |
| Gene Name | NA |
| Pfam domain motif | IP_trans |
| Motif E-value | 2.2e-54 |
| Motif start | 12 |
| Motif end | 167 |
| Protein seq | >Ps.54159.2 MMCLGRVSTHLKCITCRVIKCPYFERFNLSIETIHKADNGNSENVHGLSKEQLATREVVIIDIASTVTDYWSYAIGTSPV DLSKFKSSRTNRGPLLEGWQDLCEPVMTAYKLVTVDAPYWGFGYRLEQALLSAERALFLESHRNCFAWIDDWYGMTMEQI RELEKQSDSSLNEKLGKVASVDDTQGLDLNDSDKNMKQLS* |
| CDS seq | >Ps.54159.2 ATGATGTGTTTGGGAAGGGTCAGTACACATCTAAAGTGTATCACTTGCAGAGTGATTAAGTGCCCGTACTTCGAGAGATT CAACTTATCCATTGAAACTATTCATAAGGCTGACAATGGGAACTCAGAAAATGTGCATGGTTTAAGCAAAGAACAGCTAG CGACCAGGGAAGTAGTGATTATTGATATAGCTTCAACTGTGACGGATTATTGGAGCTATGCTATTGGTACCAGCCCTGTG GATTTGAGCAAATTCAAATCATCGAGGACAAACCGCGGTCCGCTATTGGAGGGATGGCAGGATCTGTGTGAACCAGTAAT GACAGCATATAAACTGGTGACGGTAGATGCTCCATACTGGGGTTTTGGGTATCGACTGGAACAAGCTTTACTTTCCGCTG AAAGGGCACTCTTTCTGGAAAGCCATAGGAACTGCTTTGCTTGGATTGATGATTGGTATGGGATGACAATGGAGCAAATA CGAGAACTTGAAAAACAAAGTGATTCGTCACTGAATGAGAAACTGGGGAAGGTAGCTTCAGTGGACGACACACAAGGTCT CGACTTAAATGACAGTGACAAGAATATGAAGCAACTAAGCTAG |