
| Microexon ID | Zm_7:10853791-10853797:+ |
| Species | Zea mays | Coordinates | 7:10853791..10853797 |
| Microexon Cluster ID | MEP15 |
| Size | 7 |
| Phase | 2 |
| Pfam Domain Motif | GBP |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 50,7,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | ATATCTMGRYTSTCATTTGCTGTTGARMTTGCTGAAGARTTYTATGGAAGAGTGAAGGGRCAAGATGTTGCWTTTGARCCWGCWAARCTYYTGTGGCTTATCCARMGT |
| Logo of Microexon-tag DNA Seq | 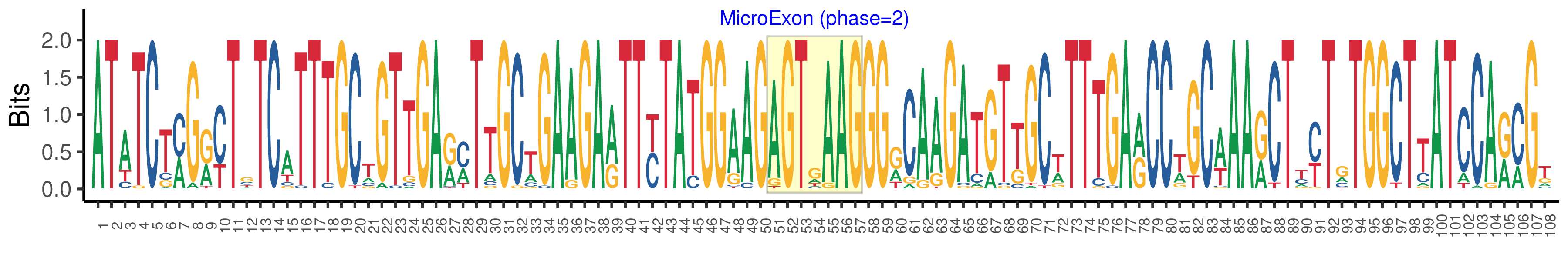 |
| Alignment of exons | 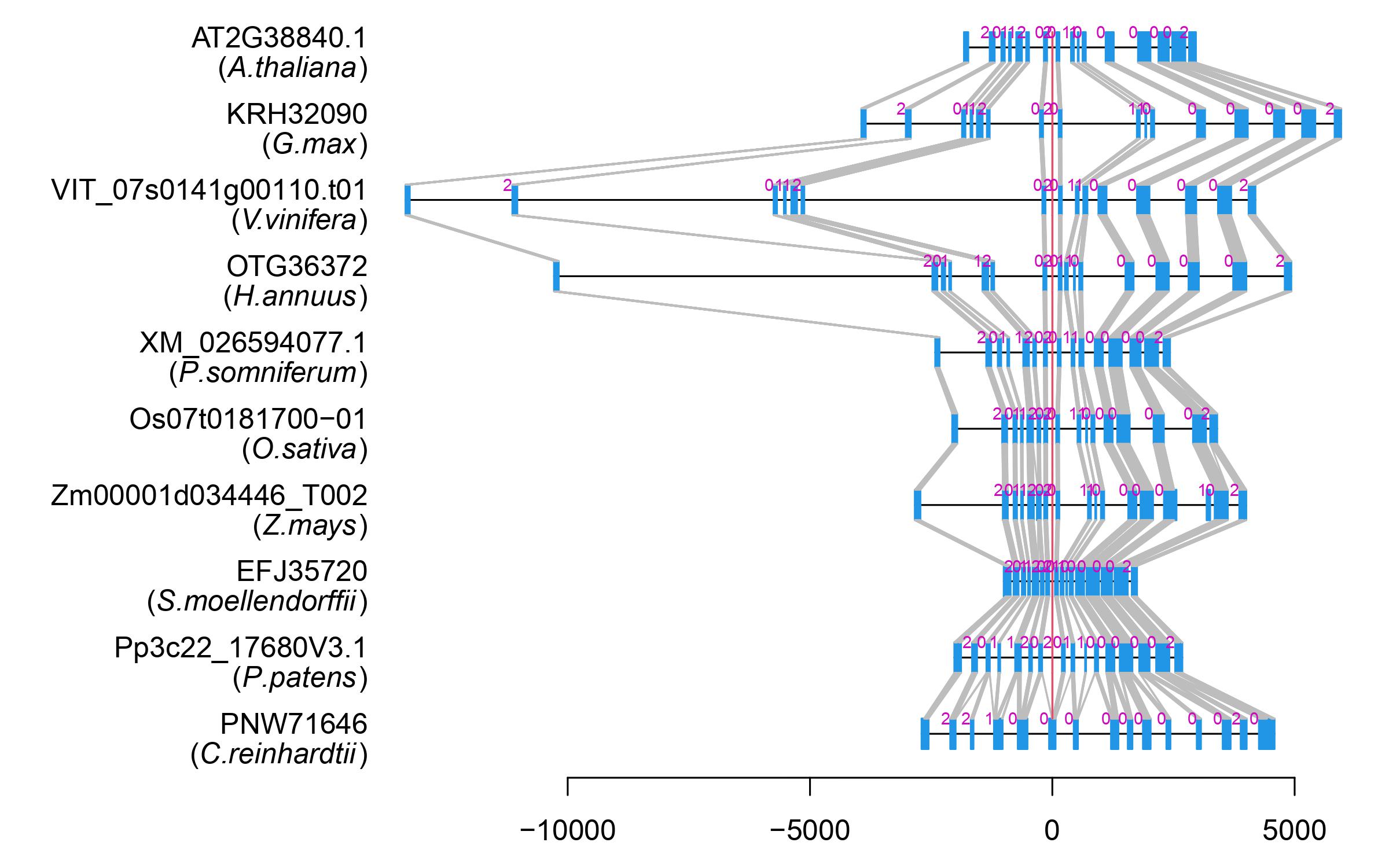 |
| Microexon DNA seq | AGTCAAG |
| Microexon Amino Acid seq | RVK |
| Microexon-tag DNA Seq | ATATCGAGACTTTCATTTGCCGTCGAAATTGCTGAAGAATTCTATGGAAGAGTCAAGGGGCAAGATGTTGCTTTTGAGCCAGCCAAACTTCTGTGGTTGATCCAGAGG |
| Microexon-tag Amino Acid Seq | ISRLSFAVEIAEEFYGRVKGQDVAFEPAKLLWLIQR |
| Microexon-tag spanning region | 10853643-10853924 |
| Microexon-tag prediction score | 0.9563 |
| Overlapped with the annotated transcript (%) | 93.52 |
| New Transcript ID | Zm00001d018964_T002x |
| Reference Transcript ID | Zm00001d018964_T002 |
| Gene ID | Zm00001d018964 |
| Gene Name | NA |
Zm_7:10853791-10853797:+ does not have available information here.
| Microexon DNA seq | AGTCAAG |
| Microexon Amino Acid seq | RVK |
| Microexon-tag DNA Seq | ATATCGAGACTTTCATTTGCCGTCGAAATTGCTGAAGAATTCTATGGAAGAGTCAAGGGGCAAGATGTTGCTTTTGAGCCAGCCAAACTTCTGTGGTTGATCCAGAGG |
| Microexon-tag Amino Acid seq | ISRLSFAVEIAEEFYGRVKGQDVAFEPAKLLWLIQR |
| Transcript ID | Zm.29659.1 |
| Gene ID | Zm.29659 |
| Gene Name | NA |
| Pfam domain motif | GBP |
| Motif E-value | 1.5e-38 |
| Motif start | 42 |
| Motif end | 282 |
| Protein seq | >Zm.29659.1 MGCAPVVWAAVWLLAAAVAGFADPGGLERAFPIVKPDYGHTKLQLAKEGLEAIQRIRTPIAAVSVIGPYRSGKSFLLNQL LSLSCDNGFGVGHMRDTKTKGIWVWGSPVEVDIGGSKVSVLYLDTEGFESVGKSNVYDDRIFALATVLSSVLIYNLPETI READISRLSFAVEIAEEFYGRVKGQDVAFEPAKLLWLIQRDFLQGKSVQQMVDEALQRVPNNNGDTNIDELNQIRDSLAV MGDNSTAFSLPQPHLQRTKLCDMEDQELDPLYVKKRDQLKEVVASMIKPKIVQGRTLNGTEFVSFLGQIVDALNKGEIPS AGSLVEIFNKAILERCLKVYSEIMGRAGLPVSVDELREFHDLAKDEARRLFNKQHFGKHHAARSILKLDEEIKKVYRNLG QANEYQSSKLCEARFSECEDKMERLQVLKLPSMAKFDAGFLLCNRSFETDCVGPAKESYRHRMSKMLARSRALFIKEYNN KLFNWLAIFSLAMVVVGRFLVKFFLLEVAAWVMFAFLETYTRLFWSSESLYYNPAWHAIVSSWETVVYSPVLDLDRWAFP ICAVLLFLAFYWRCLGGRRGIARSLLPLYNGAYYRSSSRTRTD* |
| CDS seq | >Zm.29659.1 ATGGGCTGTGCCCCGGTGGTGTGGGCCGCGGTGTGGTTGCTTGCTGCCGCGGTGGCAGGGTTCGCCGATCCCGGCGGCCT CGAGCGCGCGTTTCCAATAGTGAAGCCAGACTACGGGCACACGAAACTCCAACTCGCAAAAGAAGGTCTGGAAGCAATTC AGAGGATCAGAACTCCAATAGCTGCTGTTTCTGTTATTGGCCCCTATCGGTCGGGCAAATCTTTTCTTCTCAACCAACTT CTCTCCCTATCATGTGACAATGGTTTTGGAGTTGGACACATGCGGGATACAAAAACAAAAGGTATATGGGTATGGGGTTC CCCCGTTGAGGTGGATATCGGTGGCTCCAAGGTGTCTGTTCTATACCTGGACACCGAAGGATTTGAGAGTGTTGGAAAAT CAAATGTATATGATGATAGGATATTTGCTCTGGCAACTGTTTTAAGTTCTGTTCTGATCTACAATCTTCCTGAAACAATT CGTGAAGCCGATATATCGAGACTTTCATTTGCCGTCGAAATTGCTGAAGAATTCTATGGAAGAGTCAAGGGGCAAGATGT TGCTTTTGAGCCAGCCAAACTTCTGTGGTTGATCCAGAGGGATTTTCTCCAAGGAAAATCGGTTCAGCAGATGGTTGATG AAGCTCTCCAACGGGTACCTAACAACAATGGAGACACAAATATTGACGAGCTCAACCAAATCAGAGACTCTTTGGCGGTT ATGGGAGATAACAGTACAGCTTTTAGCTTACCACAGCCTCATCTGCAAAGGACAAAGTTATGTGATATGGAGGACCAGGA ACTTGATCCGTTGTATGTAAAGAAGAGGGATCAGTTGAAAGAAGTCGTTGCATCCATGATAAAACCAAAAATTGTGCAGG GTAGAACTCTGAATGGAACAGAGTTTGTATCCTTCCTAGGGCAGATAGTTGATGCTTTGAATAAAGGTGAAATTCCATCA GCCGGGTCGCTGGTTGAAATTTTCAATAAGGCCATTCTTGAACGTTGCTTGAAGGTGTACAGTGAAATAATGGGCAGAGC AGGTCTACCAGTGTCAGTGGATGAACTTCGGGAGTTCCACGACCTGGCAAAAGATGAAGCTAGAAGGCTTTTCAACAAGC AGCATTTCGGTAAACATCATGCTGCTCGGTCCATCCTCAAGCTTGATGAAGAGATTAAAAAGGTGTACAGAAACTTGGGC CAAGCTAACGAGTATCAGTCATCAAAGCTGTGCGAAGCACGGTTCTCAGAGTGCGAAGATAAAATGGAACGACTCCAAGT CTTGAAGCTTCCGTCCATGGCGAAATTTGATGCCGGATTCCTTCTCTGCAATCGAAGTTTTGAAACGGACTGTGTGGGGC CAGCCAAGGAAAGCTATCGACACCGGATGTCAAAGATGCTGGCAAGGTCCCGTGCTCTTTTCATCAAGGAGTACAACAAC AAGCTCTTCAACTGGCTCGCCATATTCTCCCTGGCCATGGTTGTCGTTGGGCGCTTCTTGGTCAAGTTCTTCCTGCTCGA AGTTGCTGCGTGGGTGATGTTTGCCTTCCTAGAGACATACACGAGGCTGTTCTGGTCGTCGGAGTCCCTGTACTACAACC CCGCCTGGCACGCGATCGTCTCTTCTTGGGAAACCGTCGTCTACAGCCCCGTTCTTGATCTTGACAGATGGGCGTTCCCG ATCTGCGCTGTCCTGTTGTTTTTGGCGTTTTATTGGCGTTGCCTCGGTGGAAGGAGGGGAATTGCCCGGTCACTGCTACC TCTGTACAACGGGGCTTATTACAGAAGCTCCAGTCGCACAAGAACAGACTGA |