
| Microexon ID | Zm_5:4374191-4374197:- |
| Species | Zea mays | Coordinates | 5:4374191..4374197 |
| Microexon Cluster ID | MEP15 |
| Size | 7 |
| Phase | 2 |
| Pfam Domain Motif | GBP |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 50,7,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | ATATCTMGRYTSTCATTTGCTGTTGARMTTGCTGAAGARTTYTATGGAAGAGTGAAGGGRCAAGATGTTGCWTTTGARCCWGCWAARCTYYTGTGGCTTATCCARMGT |
| Logo of Microexon-tag DNA Seq | 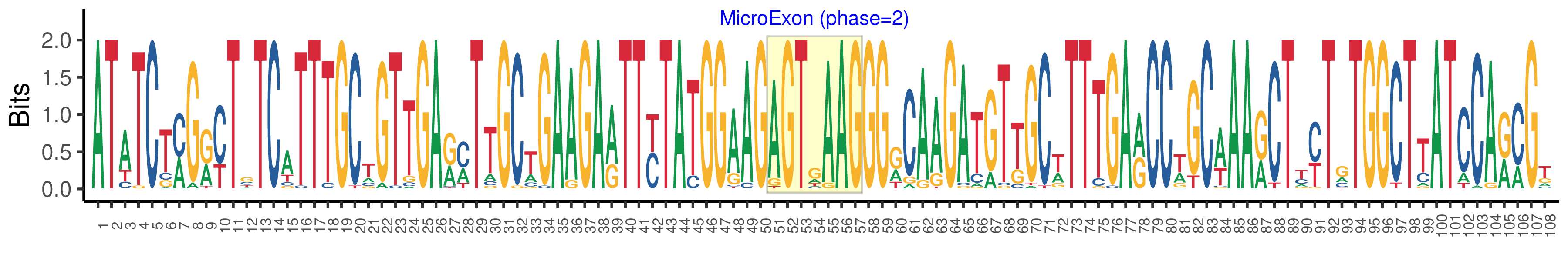 |
| Alignment of exons |  |
| Microexon DNA seq | AGTGAAG |
| Microexon Amino Acid seq | RVK |
| Microexon-tag DNA Seq | ATATCTAGGCTTTCATTTGCCGTTGAAATTGCTGAAGAGTTCTATGGAAGAGTGAAGGGGCAAGATGTTGCTTTTGAGCCAGCAAAGCTTCTCTGGCTTATCCAGAGG |
| Microexon-tag Amino Acid Seq | ISRLSFAVEIAEEFYGRVKGQDVAFEPAKLLWLIQR |
| Microexon-tag spanning region | 4374061-4374350 |
| Microexon-tag prediction score | 0.9841 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Zm00001d013075_T001x |
| Reference Transcript ID | Zm00001d013075_T001 |
| Gene ID | Zm00001d013075 |
| Gene Name | NA |
| Transcript ID | Zm00001d013075_T001 |
| Protein ID | Zm00001d013075_P001 |
| Gene ID | Zm00001d013075 |
| Gene Name | NA |
| Pfam domain motif | GBP |
| Motif E-value | 2.8e-39 |
| Motif start | 51 |
| Motif end | 297 |
| Protein seq | >Zm00001d013075_P001 MWRGRCAPGISVAALALLAVAAAAGAAAGDPDPDDLERAFPIVEPDHRHTKLRLSEQGLEAIRRIETPIAVVGVIGPYRS GKSFLLNQLLSLSCDKGFGVGHMRDTKTKGIWIWGTPVEMDVDGSKVSVLYLDTEGFESVGKSNVYDDRIFALATVLSSV LIYNLPETVREADISRLSFAVEIAEEFYGRVKGQDVAFEPAKLLWLIQRDFLQGKSVQQMVNEALQRVPNDNGDRYINEV NQIRDSLAVMGNHSTAFSLPQPHLQRTKLCDMEDKELEPLYVKRREQLKQLVSSIVKPKIVQGRTLNGKDFVSFLQQILE ALNKGEIPSTGSLVEIFNKAILDRCLKVYREKMDSLNLPVPVDELQKLHEMTNGEARILFDKQHFGKHHAAQSALKLEDE IKEVYRNFLLANEYQSSKLCEARFSECEDKMDHLQVLKLPSMAKFNAGFTHCNQSFVKDCVGPAKESYERRMSKMLAKSR ALFIKEYNNKLFNWLATFSLVMVVIGRFVIKFFLLEIVAWVMFIFLETYTRMFWSAESLYYNPAWHIIVSSWEMIVYSPI LDLDRWAIPIVIMLSLGVLYWRCFGRRRKRVKGSLLPLYKNSYKNSSRPRSD* |
| CDS seq | >Zm00001d013075_T001 ATGTGGCGGGGGCGGTGTGCCCCGGGGATCAGCGTGGCGGCGCTGGCCCTGCTCGCCGTCGCGGCTGCTGCAGGAGCCGC CGCGGGGGATCCCGATCCCGACGACCTCGAGCGCGCGTTTCCAATAGTGGAGCCAGACCACAGACACACTAAACTTCGTC TCTCAGAGCAAGGTCTAGAAGCAATTCGAAGGATCGAAACTCCTATTGCTGTTGTTGGTGTCATTGGTCCCTATCGATCT GGAAAATCATTTCTCCTCAACCAGCTTCTCTCCTTATCTTGTGATAAAGGTTTTGGAGTTGGTCATATGCGAGATACTAA AACTAAAGGTATTTGGATTTGGGGTACTCCTGTTGAGATGGATGTTGATGGTTCCAAAGTTTCTGTCCTTTATCTAGACA CTGAAGGGTTTGAAAGTGTTGGGAAATCAAATGTGTATGATGATAGGATATTTGCTTTGGCGACTGTCTTAAGCTCGGTC CTTATCTACAACCTTCCTGAGACGGTACGTGAAGCTGATATATCTAGGCTTTCATTTGCCGTTGAAATTGCTGAAGAGTT CTATGGAAGAGTGAAGGGGCAAGATGTTGCTTTTGAGCCAGCAAAGCTTCTCTGGCTTATCCAGAGGGATTTCCTCCAAG GAAAATCTGTTCAACAAATGGTCAATGAAGCCCTCCAACGGGTGCCTAATGATAATGGTGACAGATATATTAATGAGGTC AATCAAATACGAGATTCTTTGGCAGTTATGGGCAACCACAGCACTGCTTTTAGCTTGCCCCAGCCACATCTTCAAAGAAC AAAATTATGTGATATGGAGGACAAAGAGCTTGAACCCTTATATGTAAAACGGCGGGAGCAACTAAAGCAGCTTGTTTCGT CCATTGTAAAACCAAAAATCGTGCAGGGTAGAACTCTAAATGGGAAGGACTTTGTGTCTTTCTTGCAGCAGATTCTTGAG GCTTTGAATAAAGGCGAAATTCCATCAACTGGATCTCTGGTTGAAATTTTTAACAAAGCCATTCTTGATCGCTGTCTGAA GGTGTACAGGGAGAAAATGGATAGCTTAAACCTACCAGTGCCGGTAGACGAACTGCAGAAACTTCATGAGATGACAAATG GTGAAGCTAGAATTCTCTTTGATAAGCAGCACTTTGGCAAACATCATGCTGCTCAGTCGGCCCTCAAGCTTGAAGATGAG ATTAAAGAGGTGTACAGAAACTTCCTTCTAGCTAATGAATATCAATCGTCAAAGCTATGTGAAGCTCGCTTTTCTGAGTG TGAAGATAAGATGGATCACCTTCAAGTCTTGAAACTTCCTTCAATGGCAAAATTCAATGCAGGCTTTACTCACTGCAACC AAAGTTTCGTAAAGGACTGCGTGGGACCTGCAAAAGAAAGCTACGAACGTAGAATGTCAAAGATGCTTGCCAAGTCTCGT GCTCTTTTCATCAAGGAGTACAATAACAAGCTTTTCAATTGGTTGGCGACATTCTCTTTGGTCATGGTGGTGATCGGGCG CTTTGTCATCAAGTTCTTCCTACTTGAAATTGTTGCTTGGGTGATGTTTATCTTCCTAGAGACGTACACGAGAATGTTCT GGTCAGCGGAATCACTGTATTATAATCCAGCTTGGCACATCATTGTTTCTTCCTGGGAGATGATCGTGTACAGCCCTATT CTTGATCTGGACAGATGGGCCATCCCAATTGTCATAATGCTATCATTGGGGGTTCTTTACTGGAGGTGCTTCGGCCGCAG AAGGAAGCGGGTAAAGGGTTCCCTCCTCCCATTGTACAAGAATTCTTACAAGAACTCGAGTCGCCCGAGATCAGATTAG |
| Microexon DNA seq | AGTGAAG |
| Microexon Amino Acid seq | RVK |
| Microexon-tag DNA Seq | ATATCTAGGCTTTCATTTGCCGTTGAAATTGCTGAAGAGTTCTATGGAAGAGTGAAGGGGCAAGATGTTGCTTTTGAGCCAGCAAAGCTTCTCTGGCTTATCCAGAGG |
| Microexon-tag Amino Acid seq | ISRLSFAVEIAEEFYGRVKGQDVAFEPAKLLWLIQR |
| Transcript ID | Zm.21758.2 |
| Gene ID | Zm.21758 |
| Gene Name | NA |
| Pfam domain motif | GBP |
| Motif E-value | 1.1e-39 |
| Motif start | 51 |
| Motif end | 297 |
| Protein seq | >Zm.21758.2 MWRGRCAPGISVAALALLAVAAAAGAAAGDPDPDDLERAFPIVEPDHRHTKLRLSEQGLEAIRRIETPIAVVGVIGPYRS GKSFLLNQLLSLSCDKGFGVGHMRDTKTKGIWIWGTPVEMDVDGSKVSVLYLDTEGFESVGKSNVYDDRIFALATVLSSV LIYNLPETVREADISRLSFAVEIAEEFYGRVKGQDVAFEPAKLLWLIQRDFLQGKSVQQMVNEALQRVPNDNGDRYINEV NQIRDSLAVMGNHSTAFSLPQPHLQRTKLCDMEDKELEPLYVKRREQLKQLVSSIVKPKIVQGRTLNGKDFVSFLQQILE ALNKGEIPSTGSLVEIFNKAILDRCLKVYREKMDSLNLPVPVDELQKLHEMTNGEARILFDKQHFGKHHAAQSALKLEDE IKEDVLCRCTETSF* |
| CDS seq | >Zm.21758.2 ATGTGGCGGGGGCGGTGTGCCCCGGGGATCAGCGTGGCGGCGCTGGCCCTGCTCGCCGTCGCGGCTGCTGCAGGAGCCGC CGCGGGGGATCCCGATCCCGACGACCTCGAGCGCGCGTTTCCAATAGTGGAGCCAGACCACAGACACACTAAACTTCGTC TCTCAGAGCAAGGTCTAGAAGCAATTCGAAGGATCGAAACTCCTATTGCTGTTGTTGGTGTCATTGGTCCCTATCGATCT GGAAAATCATTTCTCCTCAACCAGCTTCTCTCCTTATCTTGTGATAAAGGTTTTGGAGTTGGTCATATGCGAGATACTAA AACTAAAGGTATTTGGATTTGGGGTACTCCTGTTGAGATGGATGTTGATGGTTCCAAAGTTTCTGTCCTTTATCTAGACA CTGAAGGGTTTGAAAGTGTTGGGAAATCAAATGTGTATGATGATAGGATATTTGCTTTGGCGACTGTCTTAAGCTCGGTC CTTATCTACAACCTTCCTGAGACGGTACGTGAAGCTGATATATCTAGGCTTTCATTTGCCGTTGAAATTGCTGAAGAGTT CTATGGAAGAGTGAAGGGGCAAGATGTTGCTTTTGAGCCAGCAAAGCTTCTCTGGCTTATCCAGAGGGATTTCCTCCAAG GAAAATCTGTTCAACAAATGGTCAATGAAGCCCTCCAACGGGTGCCTAATGATAATGGTGACAGATATATTAATGAGGTC AATCAAATACGAGATTCTTTGGCAGTTATGGGCAACCACAGCACTGCTTTTAGCTTGCCCCAGCCACATCTTCAAAGAAC AAAATTATGTGATATGGAGGACAAAGAGCTTGAACCCTTATATGTAAAACGGCGGGAGCAACTAAAGCAGCTTGTTTCGT CCATTGTAAAACCAAAAATCGTGCAGGGTAGAACTCTAAATGGGAAGGACTTTGTGTCTTTCTTGCAGCAGATTCTTGAG GCTTTGAATAAAGGCGAAATTCCATCAACTGGATCTCTGGTTGAAATTTTTAACAAAGCCATTCTTGATCGCTGTCTGAA GGTGTACAGGGAGAAAATGGATAGCTTAAACCTACCAGTGCCGGTAGACGAACTGCAGAAACTTCATGAGATGACAAATG GTGAAGCTAGAATTCTCTTTGATAAGCAGCACTTTGGCAAACATCATGCTGCTCAGTCGGCCCTCAAGCTTGAAGATGAG ATTAAAGAGGATGTACTTTGTAGGTGTACAGAAACTTCCTTCTAG |