
| Microexon ID | Sm_GL377608:238641-238647:+ |
| Species | Selaginella moellendorffii | Coordinates | GL377608:238641..238647 |
| Microexon Cluster ID | MEP14 |
| Size | 7 |
| Phase | 1 |
| Pfam Domain Motif | Prefoldin_2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 52,7,49 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTGGAGGARCTGMRVCARYTRYCTGATGAYACAAAYACWTACAARTCYATWGGRARAAYGTTTRTTTTRGAGCCMAARTCAGTTYTRRTGRATGARCARGARMARAAR |
| Logo of Microexon-tag DNA Seq | 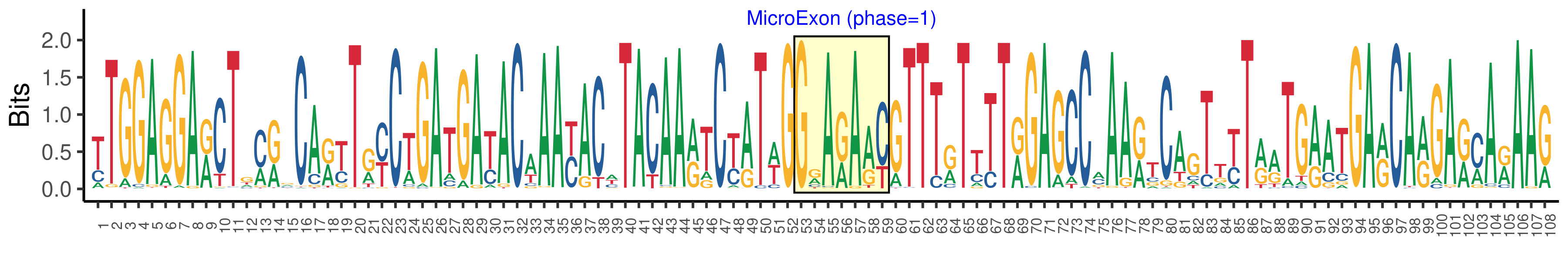 |
| Alignment of exons | 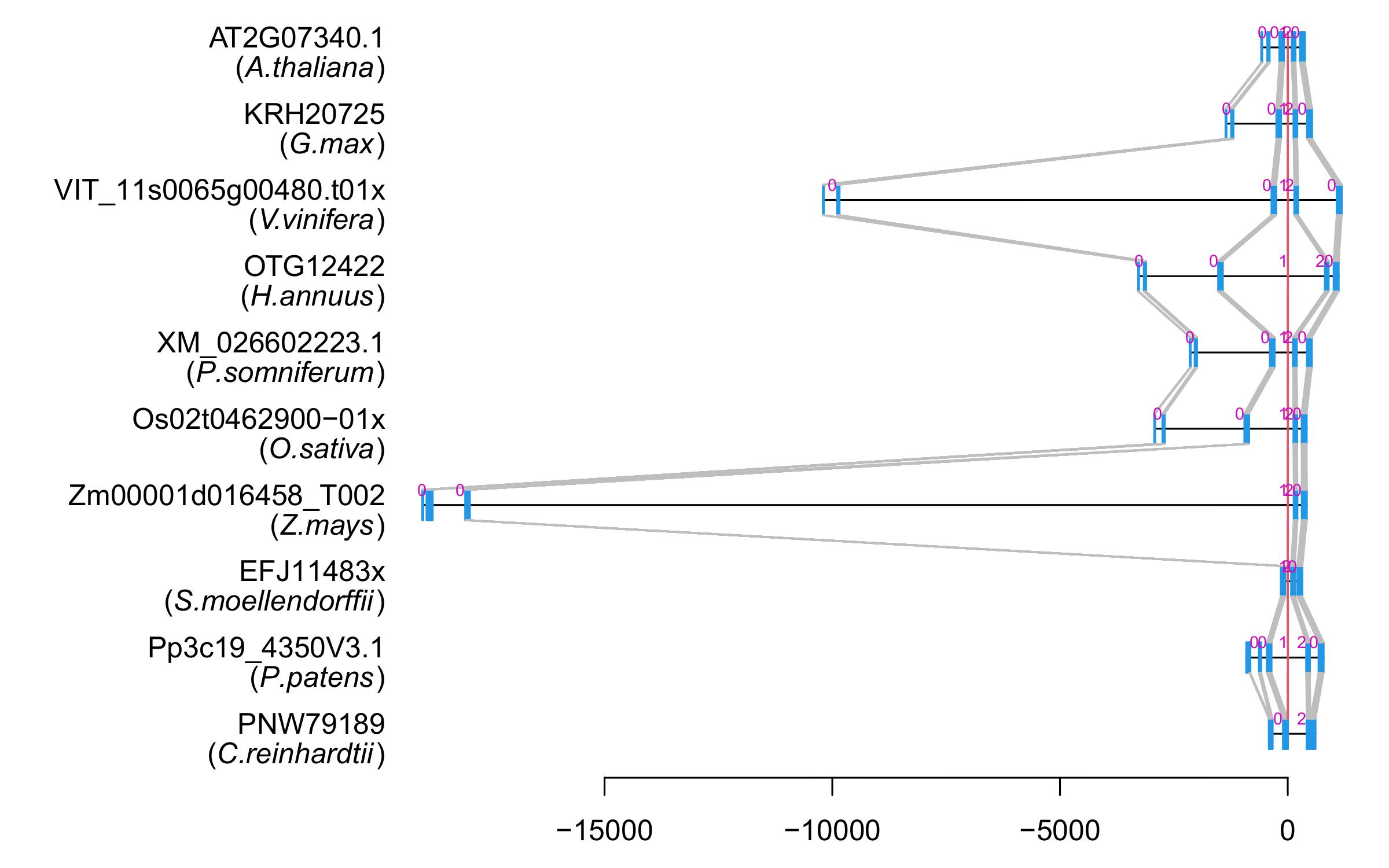 |
| Microexon DNA seq | GCAAAGC |
| Microexon Amino Acid seq | GKA |
| Microexon-tag DNA Seq | CTAGACGAGATCAACTACCTAGACGATGATACACACACTTACAAATCAGTCGGCAAAGCCTTTATTTTGGAGCCCAAGTCGAAGCTCGTGGGCGAGCTACAAGAAAAG |
| Microexon-tag Amino Acid Seq | LDEINYLDDDTHTYKSVGKAFILEPKSKLVGELQEK |
| Microexon-tag spanning region | 238536-238762 |
| Microexon-tag prediction score | 0.8525 |
| Overlapped with the annotated transcript (%) | 70.4 |
| New Transcript ID | EFJ18785x |
| Reference Transcript ID | EFJ18785 |
| Gene ID | SELMODRAFT_112137 |
| Gene Name | NA |
Sm_GL377608:238641-238647:+ does not have available information here.
| Microexon DNA seq | GCAAAGC |
| Microexon Amino Acid seq | GKA |
| Microexon-tag DNA Seq | CTAGACGAGATCAACTACCTAGACGATGATACACACACTTACAAATCAGTCGGCAAAGCCTTTATTTTGGAGCCCAAGTCGAAGCTCGTGGGCGAGCTACAAGAAAAG |
| Microexon-tag Amino Acid seq | LDEINYLDDDTHTYKSVGKAFILEPKSKLVGELQEK |
| Transcript ID | Sm.18686.3 |
| Gene ID | Sm.18686 |
| Gene Name | NA |
| Pfam domain motif | Prefoldin_2 |
| Motif E-value | 2.9e-17 |
| Motif start | 750 |
| Motif end | 853 |
| Protein seq | >Sm.18686.3 MPARLPNVLINGASGIAVGMATNIPPHNLSEVVDALCALIKNPNATVEELMEHLPGPDFPTGGQILGISGIADAFRTGRG TITVRGKADFEELEGKISRSAIVITEIPYGTNKSTLVAKIAELVNSKMLEGVSDIRDESDRAGMRIVVELKRGAVAAVVL NNLYKHTALQSRFSYNLVGILNKEPEVFSIKRLLEIFLDFRCSVVQRRAQCELTRAEARDHLLEGFLKGLDKLDDVVKVL KSAKDSASAMQALQKDFGLSAAQAEALLGMPLRRITSLERNKILEEHNSLLAQASDLRELLSNKQRVLKVVEKEALALKN EFGSPRRTVLEEYGDGRIDNIDVIPNNESFLTLSEKGYIKRMQPDTFLAQRRGTTGKSGAKMKSNDGMVDSFMCRNHDHV LVFSERGVVYSFRAYQVPECSRTSTGVPIIQVLSIPAGERITSIFPLSEFREDHFLVMLTSKGYIKRTELPAFSSIRPQG IVAIQLMEGDELKWVRLATQSDRIFIGSRNGMAMQAPCEFRSMRRGARGLRAMRIKEGDEIAAIDIVPGSVWKERSKASE APWLLFVSENGMAKRVPITSFPRHGLNKVGVIGCKFVPGDRLASMFVVGTSVSDGESNEEIVVGTQGGIFNRFKVREISI KSRMGRCFSFVSCKLLFDGLWLSLQGHQADETRCGRQGEIGLCGCSVGFSRSRRTTIRFIPGIVEEHGHYVQGLLLYSKN FGTDFIRRVYSLFPPPCAMADTMNKEAVVELQAKLVDATAKLKQVQMQMRTKETEKKRALLTLDEINYLDDDTHTYKSVG KAFILEPKSKLVGELQEKAQECESAMTNLSASKEYLERQLKEIEGNFKELLQQSPSLARQVMAMSVT* |
| CDS seq | >Sm.18686.3 ATGCCTGCCAGACTTCCCAACGTTCTTATCAATGGTGCTTCCGGAATAGCCGTTGGCATGGCTACCAATATACCTCCGCA CAACCTCTCCGAAGTTGTGGACGCACTTTGCGCTTTGATAAAGAATCCTAATGCAACTGTCGAAGAACTGATGGAACATC TGCCTGGCCCAGATTTCCCGACCGGAGGACAAATTTTGGGGATATCGGGGATAGCGGACGCTTTTAGAACAGGCCGCGGA ACGATTACAGTTAGAGGCAAAGCAGATTTCGAGGAGCTGGAAGGAAAAATTTCTCGCTCAGCTATCGTTATTACTGAGAT TCCGTATGGTACAAACAAGTCTACTTTGGTTGCTAAGATTGCCGAACTCGTTAACAGTAAGATGCTAGAAGGTGTAAGCG ACATCAGAGATGAAAGTGACCGAGCAGGAATGCGGATTGTCGTCGAGCTCAAAAGAGGCGCTGTGGCAGCTGTTGTTCTG AATAACCTGTACAAGCATACAGCCCTGCAGTCTCGTTTTAGCTACAACCTAGTTGGCATTCTTAACAAGGAGCCGGAGGT GTTCAGTATCAAGAGACTGCTGGAGATATTTCTGGATTTTAGGTGTTCAGTAGTCCAGAGGCGAGCTCAGTGCGAACTTA CACGTGCTGAAGCAAGGGATCACTTGTTGGAGGGTTTTTTGAAAGGATTGGACAAGCTGGACGATGTTGTGAAAGTGTTA AAGAGTGCCAAGGATTCTGCGAGTGCAATGCAGGCATTGCAGAAAGATTTCGGACTATCTGCAGCCCAGGCCGAAGCCCT TTTAGGAATGCCGCTACGGCGTATAACCAGTTTAGAGCGAAATAAGATACTCGAGGAACATAATAGTCTTCTTGCGCAGG CCTCCGACTTGCGTGAGTTGCTCTCAAACAAACAGCGCGTTTTAAAGGTGGTTGAAAAAGAAGCTTTGGCTTTGAAGAAC GAGTTTGGAAGTCCACGTCGGACTGTGCTCGAGGAGTATGGCGATGGTCGAATAGATAACATTGATGTGATACCAAACAA CGAGAGTTTCCTGACTCTCAGCGAGAAGGGTTACATCAAGAGGATGCAGCCCGACACCTTTTTAGCACAAAGGCGTGGGA CAACAGGCAAGTCCGGGGCCAAGATGAAAAGCAACGATGGCATGGTGGATTCATTTATGTGCAGGAATCACGACCACGTT CTCGTATTCAGCGAGCGGGGAGTTGTATACTCCTTTCGTGCCTACCAAGTCCCTGAATGCTCAAGGACCTCTACTGGTGT TCCCATCATTCAGGTGCTCTCCATCCCGGCGGGAGAACGAATTACTTCTATCTTCCCACTCAGCGAGTTCCGAGAGGACC ATTTTCTGGTCATGCTCACTTCCAAAGGTTACATCAAACGAACGGAACTACCAGCTTTCTCATCCATTCGACCCCAAGGC ATTGTGGCCATTCAGCTGATGGAGGGAGACGAGCTTAAATGGGTGCGACTGGCCACCCAATCTGACAGGATATTCATTGG CTCTCGGAATGGGATGGCTATGCAGGCTCCTTGTGAGTTTCGTTCCATGCGAAGGGGAGCTCGTGGTCTAAGGGCCATGC GCATCAAAGAAGGTGACGAGATTGCTGCTATCGACATTGTTCCTGGATCTGTATGGAAAGAGAGGAGCAAAGCAAGCGAA GCTCCTTGGTTGCTCTTCGTCTCTGAAAACGGGATGGCGAAGCGAGTCCCTATAACTTCCTTCCCTCGGCACGGCTTGAA CAAAGTTGGGGTTATCGGATGCAAGTTCGTTCCCGGGGACCGGCTCGCATCCATGTTTGTCGTGGGAACGTCAGTTTCAG ATGGAGAAAGCAACGAAGAAATCGTAGTCGGGACCCAGGGAGGGATCTTCAACCGGTTCAAAGTTCGCGAGATCAGTATC AAGTCGCGGATGGGAAGGTGCTTTTCTTTCGTTTCCTGCAAGCTTTTGTTTGATGGTCTGTGGTTGTCTCTCCAGGGGCA TCAAGCTGATGAAACTAGATGTGGACGACAAGGTGAAATCGGTCTCTGTGGTTGCAGCGTCGGATTCTCCCGCTCACGAC GAACCACCATCCGATTCATCCCAGGAATCGTAGAAGAACACGGCCATTACGTACAGGGCTTACTTTTGTATAGCAAAAAT TTTGGTACGGACTTTATAAGACGGGTATATTCTCTCTTTCCTCCTCCGTGTGCTATGGCGGATACCATGAACAAGGAGGC TGTTGTGGAGCTCCAGGCCAAGCTGGTGGATGCCACAGCCAAGCTCAAGCAGGTCCAAATGCAAATGCGGACCAAAGAAA CCGAGAAGAAGCGCGCTCTGTTGACACTAGACGAGATCAACTACCTAGACGATGATACACACACTTACAAATCAGTCGGC AAAGCCTTTATTTTGGAGCCCAAGTCGAAGCTCGTGGGCGAGCTACAAGAAAAGGCCCAGGAATGCGAGTCCGCCATGAC CAATCTCTCCGCTTCCAAGGAGTATCTAGAACGGCAGCTCAAAGAAATCGAGGGTAATTTCAAGGAACTGTTGCAGCAAT CACCATCCCTCGCAAGACAAGTCATGGCAATGTCAGTGACATAA |