
| Microexon ID | Sb_4:39531694-39531700:+ |
| Species | Sorghum Bicolor | Coordinates | 4:39531694..39531700 |
| Microexon Cluster ID | MEP14 |
| Size | 7 |
| Phase | 1 |
| Pfam Domain Motif | Prefoldin_2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 52,7,49 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTGGAGGARCTGMRVCARYTRYCTGATGAYACAAAYACWTACAARTCYATWGGRARAAYGTTTRTTTTRGAGCCMAARTCAGTTYTRRTGRATGARCARGARMARAAR |
| Logo of Microexon-tag DNA Seq | 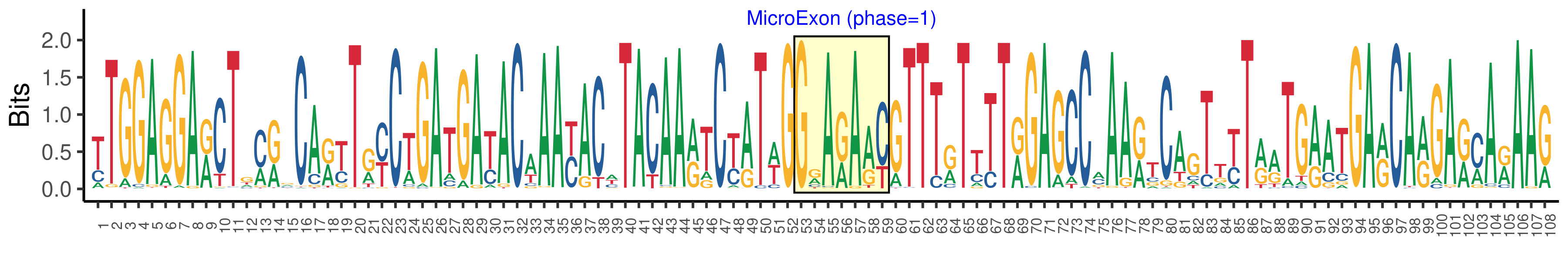 |
| Alignment of exons | 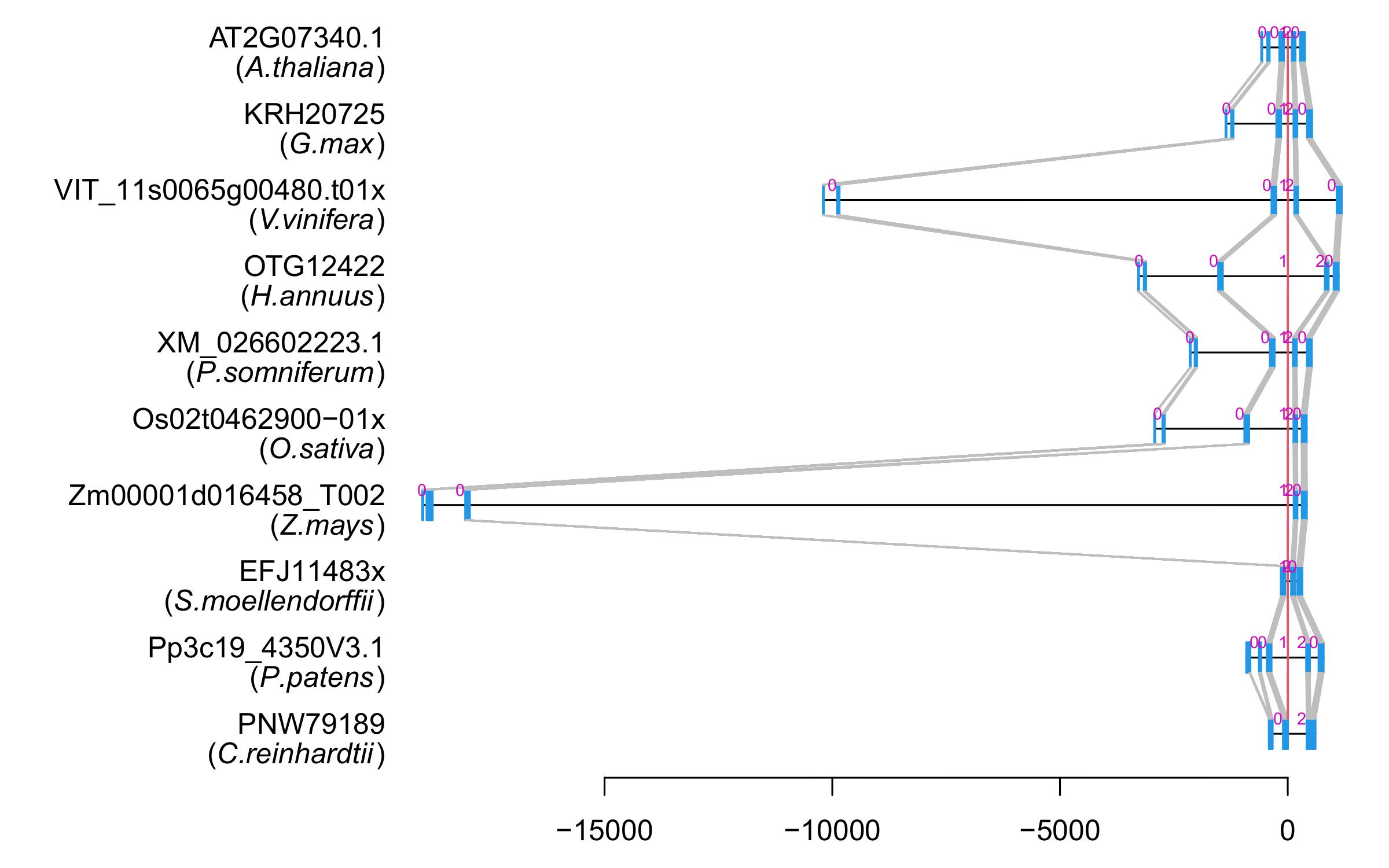 |
| Microexon DNA seq | GAAAAGT |
| Microexon Amino Acid seq | GKV |
| Microexon-tag DNA Seq | TTGGAGGAACTTCGCCAGTTGCCAGATGATACCAACACCTACAAGACTGTCGGAAAAGTGTTTATTCTGGAGCCAAAATCAGTTCTGTTTAATGAGCAGGAGCAAAAG |
| Microexon-tag Amino Acid Seq | MEELRQLPDDTNTYKTVGKVFILEPKSVLFNEQEQK |
| Microexon-tag spanning region | 39530486-39531868 |
| Microexon-tag prediction score | 0.9631 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | EES05005x |
| Reference Transcript ID | EES05005 |
| Gene ID | SORBI_3004G138800 |
| Gene Name | NA |
| Transcript ID | EES05005 |
| Protein ID | |
| Gene ID | SORBI_3004G138800 |
| Gene Name | |
| Pfam domain motif | Prefoldin_2 |
| Motif E-value | 5.20E-18 |
| Motif start | 12 |
| Motif end | 115 |
| Protein seq | >EES05005 MADEANRAAFMELQSRMIDTTAKIKQLQTQMRSKEGEKKRAYLTLEELRQLPDDTNTYKTVGKVFILEPKSVLFNEQEQK LHDSESAIASMQTSKEYLEKQQGEVENNIRELLQQDPGLARQILSMTVQ* |
| CDS seq | >EES05005 ATGGCAGACGAAGCCAACCGAGCGGCCTTCATGGAGCTGCAGTCCCGGATGATCGATACCACCGCGAAGATCAAGCAGTT ACAAACCCAAATGCGTTCTAAGGAAGGAGAAAAGAAGCGTGCCTACCTTACTTTGGAGGAACTTCGCCAGTTGCCAGATG ATACCAACACCTACAAGACTGTCGGAAAAGTGTTTATTCTGGAGCCAAAATCAGTTCTGTTTAATGAGCAGGAGCAAAAG CTTCATGACAGTGAGAGTGCTATAGCTTCAATGCAGACATCCAAAGAATACCTTGAGAAACAGCAGGGCGAAGTGGAGAA CAACATTAGGGAGCTGCTTCAACAGGATCCAGGGCTTGCGCGTCAGATCCTCTCAATGACCGTTCAATGA |
Sb_4:39531694-39531700:+ does not have available information here.