
| Microexon ID | Ha_12:14142501-14142507:- |
| Species | Helianthus annuus | Coordinates | 12:14142501..14142507 |
| Microexon Cluster ID | MEP14 |
| Size | 7 |
| Phase | 1 |
| Pfam Domain Motif | Prefoldin_2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 52,7,49 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTGGAGGARCTGMRVCARYTRYCTGATGAYACAAAYACWTACAARTCYATWGGRARAAYGTTTRTTTTRGAGCCMAARTCAGTTYTRRTGRATGARCARGARMARAAR |
| Logo of Microexon-tag DNA Seq | 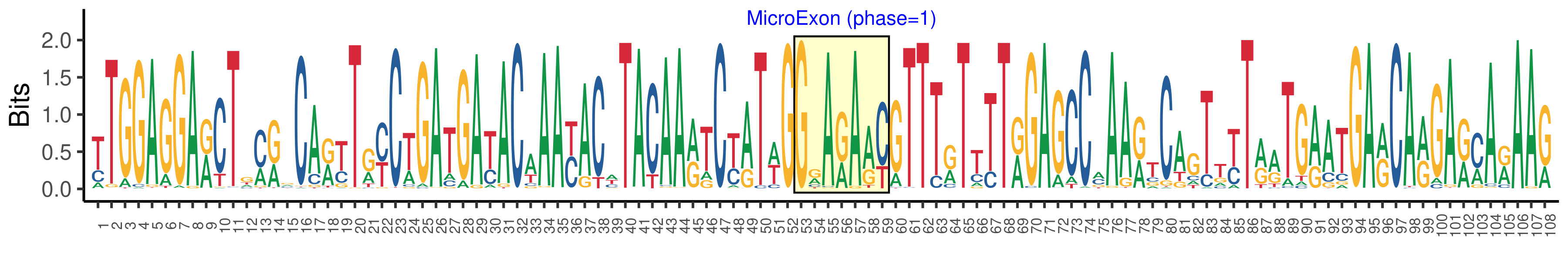 |
| Alignment of exons | 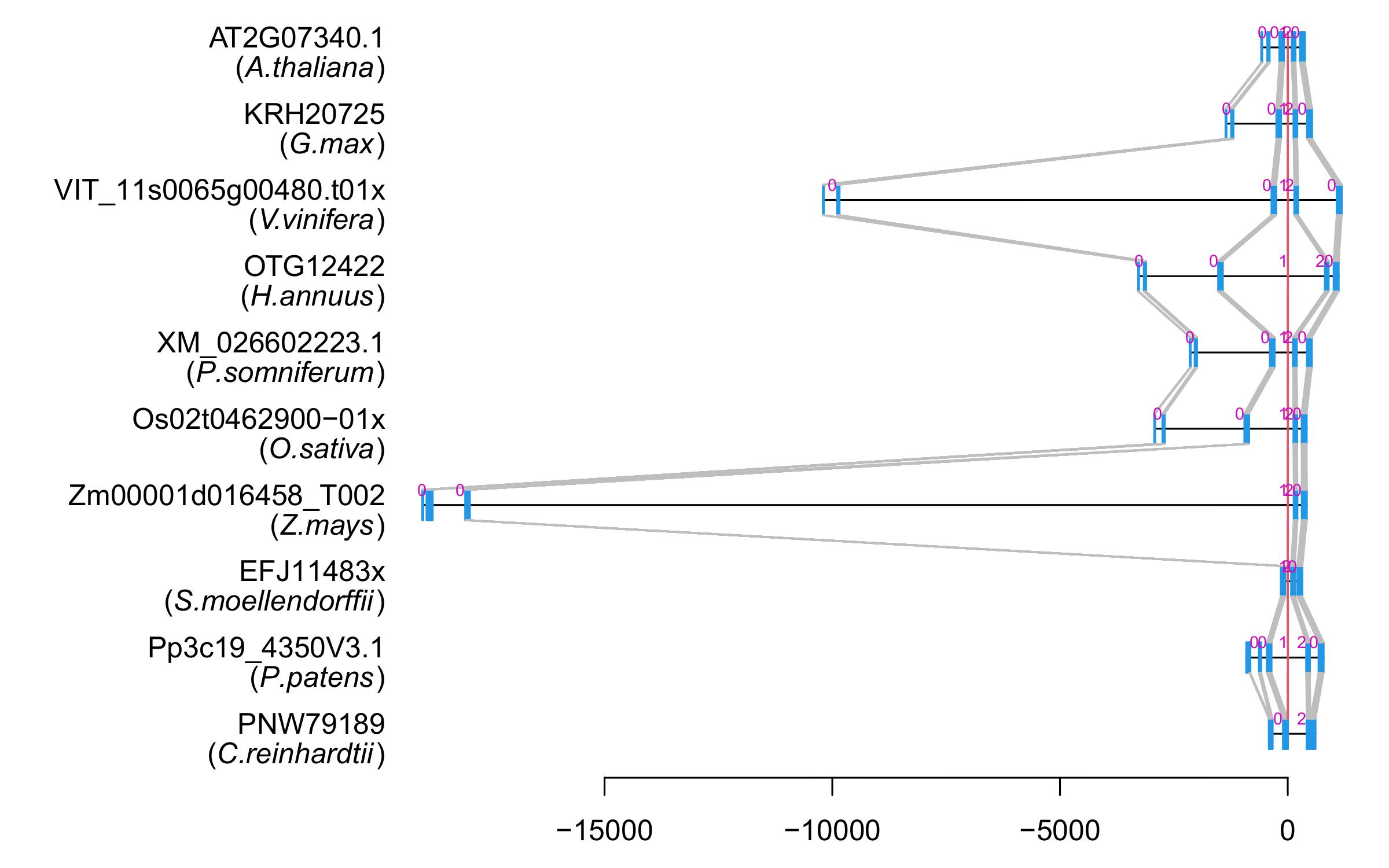 |
| Microexon DNA seq | GGAGAAC |
| Microexon Amino Acid seq | GRT |
| Microexon-tag DNA Seq | TTGGAGGAACTGCGCCAATTATCTGATGACACAAACACATACAAATCTATAGGGAGAACATTCATCTTAGAGCCAAAACCATTTCTAATGAATGAACAAGAAAAGAAG |
| Microexon-tag Amino Acid Seq | MEELRQLSDDTNTYKSIGRTFILEPKPFLMNEQEKK |
| Microexon-tag spanning region | 14141418-14142688 |
| Microexon-tag prediction score | 0.9705 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | OTG04099x |
| Reference Transcript ID | OTG04099 |
| Gene ID | HannXRQ_Chr12g0358571 |
| Gene Name | PFD1 |
| Transcript ID | OTG04099 |
| Protein ID | OTG04099 |
| Gene ID | HannXRQ_Chr12g0358571 |
| Gene Name | PFD1 |
| Pfam domain motif | Prefoldin_2 |
| Motif E-value | 7.9e-20 |
| Motif start | 13 |
| Motif end | 115 |
| Protein seq | >OTG04099 MADEANKTAFVEIQGRMIETTAKLKQVQNQMRNKESEKKRAYLTLEELRQLSDDTNTYKSIGRTFILEPKPFLMNEQEKK LSDSETAIASLQTSKEYLEKQLAEVETNLRELLQQDPSLARQIMSMAVV* |
| CDS seq | >OTG04099 ATGGCCGACGAAGCAAACAAAACAGCGTTTGTGGAGATTCAAGGTCGCATGATTGAAACAACTGCTAAATTGAAACAAGT TCAAAACCAGATGCGAAACAAAGAAAGTGAAAAGAAACGTGCTTATTTAACCTTGGAGGAACTGCGCCAATTATCTGATG ACACAAACACATACAAATCTATAGGGAGAACATTCATCTTAGAGCCAAAACCATTTCTAATGAATGAACAAGAAAAGAAG CTTAGTGATAGTGAAACTGCAATTGCCTCCTTACAGACTTCAAAGGAGTACCTTGAGAAACAATTGGCGGAGGTGGAGAC TAACTTGAGGGAGCTTTTGCAACAAGATCCCAGTCTTGCTCGTCAAATAATGTCAATGGCTGTCGTGTAA |
| Microexon DNA seq | GGAGAAC |
| Microexon Amino Acid seq | GRT |
| Microexon-tag DNA Seq | TTGGAGGAACTGCGCCAATTATCTGATGACACAAACACATACAAATCTATAGGGAGAACATTCATCTTAGAGCCAAAACCATTTCTAATGAATGAACAAGAAAAGAAG |
| Microexon-tag Amino Acid seq | MEELRQLSDDTNTYKSIGRTFILEPKPFLMNEQEKK |
| Transcript ID | OTG04099 |
| Gene ID | Ha.11405 |
| Gene Name | PFD1 |
| Pfam domain motif | Prefoldin_2 |
| Motif E-value | 7.9e-20 |
| Motif start | 13 |
| Motif end | 115 |
| Protein seq | >OTG04099 MADEANKTAFVEIQGRMIETTAKLKQVQNQMRNKESEKKRAYLTLEELRQLSDDTNTYKSIGRTFILEPKPFLMNEQEKK LSDSETAIASLQTSKEYLEKQLAEVETNLRELLQQDPSLARQIMSMAVV* |
| CDS seq | >OTG04099 ATGGCCGACGAAGCAAACAAAACAGCGTTTGTGGAGATTCAAGGTCGCATGATTGAAACAACTGCTAAATTGAAACAAGT TCAAAACCAGATGCGAAACAAAGAAAGTGAAAAGAAACGTGCTTATTTAACCTTGGAGGAACTGCGCCAATTATCTGATG ACACAAACACATACAAATCTATAGGGAGAACATTCATCTTAGAGCCAAAACCATTTCTAATGAATGAACAAGAAAAGAAG CTTAGTGATAGTGAAACTGCAATTGCCTCCTTACAGACTTCAAAGGAGTACCTTGAGAAACAATTGGCGGAGGTGGAGAC TAACTTGAGGGAGCTTTTGCAACAAGATCCCAGTCTTGCTCGTCAAATAATGTCAATGGCTGTCGTGTAA |