
| Microexon ID | Vv_13:4754271-4754276:+ |
| Species | Vistis vinifera | Coordinates | 13:4754271..4754276 |
| Microexon Cluster ID | MEP10 |
| Size | 6 |
| Phase | 0 |
| Pfam Domain Motif | DUF4788 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 51,6,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTTGARGAYTAYATYGADCCMCTYAAGRTKTACCTGRMTAGRTACAGAGAGWTGGAGGGTGAYACYAAGGGATCTGCWARRGSTGGWGATGSATCTGCTAARARRGAT |
| Logo of Microexon-tag DNA Seq | 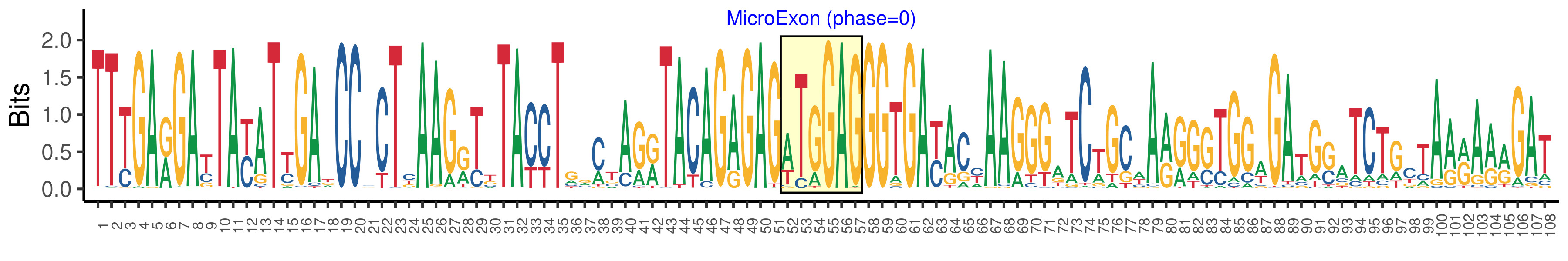 |
| Alignment of exons | 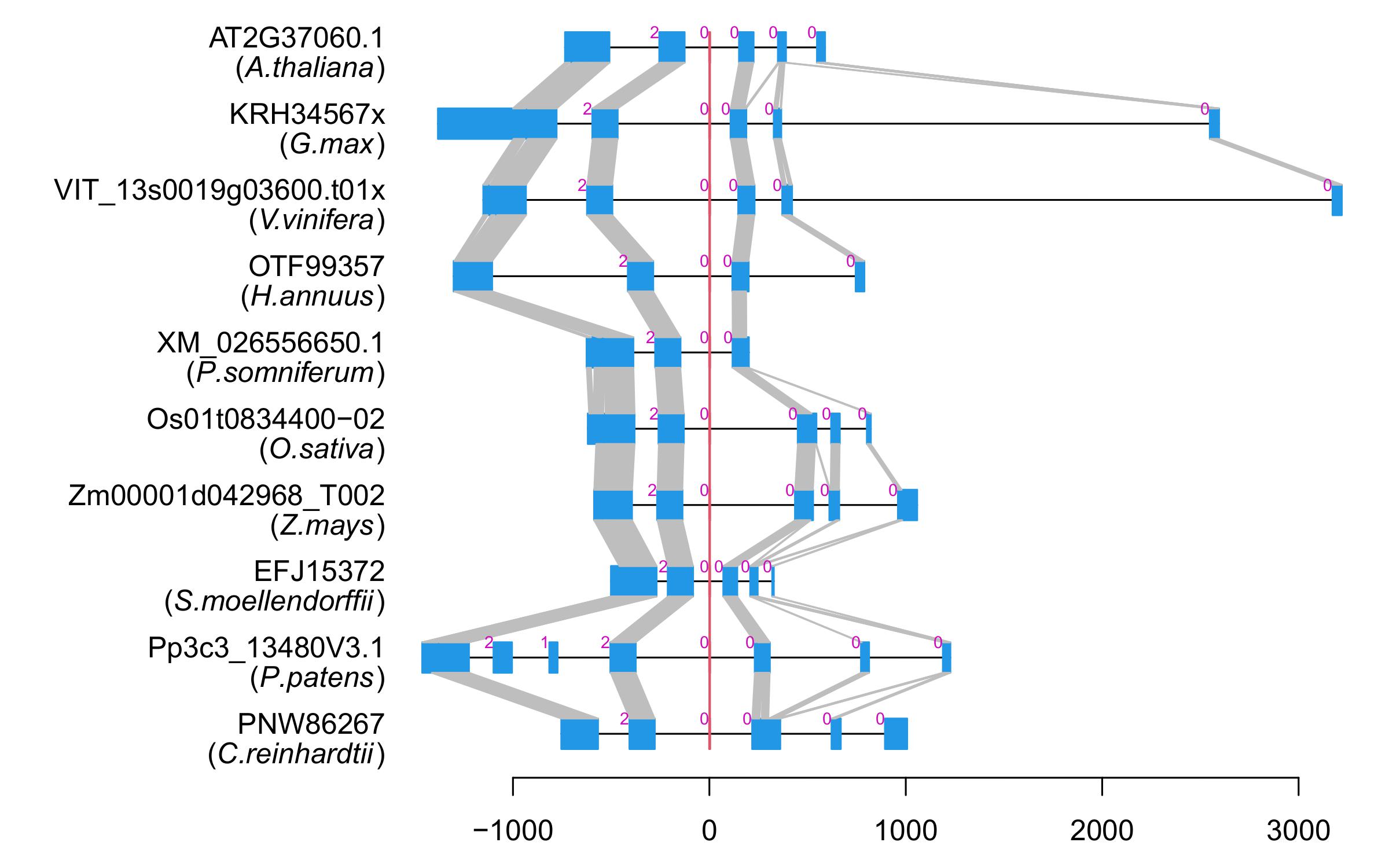 |
| Microexon DNA seq | CTGGAG |
| Microexon Amino Acid seq | LE |
| Microexon-tag DNA Seq | TTTGAAGATTATATTGAACCACTAAAGGTGTACCTGCAGAGATATAGAGAGCTGGAGGGTGATACCAGGGGATCTGCTAGAGGTGGAGATGGATCTGCTAGAAGGGAT |
| Microexon-tag Amino Acid Seq | FEDYIEPLKVYLQRYRELEGDTRGSARGGDGSARRD |
| Microexon-tag spanning region | 4753728-4754469 |
| Microexon-tag prediction score | 0.9615 |
| Overlapped with the annotated transcript (%) | 94.44 |
| New Transcript ID | VIT_13s0019g03600.t01x |
| Reference Transcript ID | VIT_13s0019g03600.t01 |
| Gene ID | VIT_13s0019g03600 |
| Gene Name | NA |
Vv_13:4754271-4754276:+ does not have available information here.
| Microexon DNA seq | CTGGAG |
| Microexon Amino Acid seq | LE |
| Microexon-tag DNA Seq | TTTGAAGATTATATTGAACCACTAAAGGTGTACCTGCAGAGATATAGAGAGCTGGAGGGTGATACCAGGGGATCTGCTAGAGGTGGAGATGGATCTGCTAGAAGGGAT |
| Microexon-tag Amino Acid seq | FEDYIEPLKVYLQRYRELEGDTRGSARGGDGSARRD |
| Transcript ID | Vv.6991.2 |
| Gene ID | Vv.6991 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >Vv.6991.2 MADGPASPPGGSHESGGDQSPRHNVREQDRYLPIANISRIMKKALPANGKIAKDAKDTVQECVSEFISFITSEASDKCQK EKRKTINGDDLLWAMATLGFEDYIEPLKVYLQRYRELEGDTRGSARGGDGSARRDAIGSQPGPNAQFAHQGSFTQAMNYM NSQAQGQHLIVSPVQSSE* |
| CDS seq | >Vv.6991.2 ATGGCAGATGGTCCGGCGAGTCCCCCCGGAGGCAGCCACGAAAGCGGCGGGGACCAGAGCCCCCGCCACAACGTTCGTGA GCAGGACCGGTACCTTCCGATCGCCAACATCAGCCGCATCATGAAGAAGGCGTTGCCGGCTAATGGTAAGATCGCCAAGG ATGCGAAGGACACTGTCCAAGAGTGCGTCTCCGAGTTCATCAGTTTCATCACCAGCGAGGCTAGTGATAAGTGTCAGAAA GAGAAGCGAAAGACGATTAATGGTGATGATCTGCTTTGGGCGATGGCGACCCTAGGATTTGAAGATTATATTGAACCACT AAAGGTGTACCTGCAGAGATATAGAGAGCTGGAGGGTGATACCAGGGGATCTGCTAGAGGTGGAGATGGATCTGCTAGAA GGGATGCAATAGGAAGTCAACCTGGTCCAAATGCACAGTTTGCTCATCAGGGGTCCTTTACGCAAGCAATGAACTATATG AATTCCCAGGCACAGGGGCAGCATCTAATTGTATCTCCCGTGCAAAGTAGCGAGTAG |