
| Microexon ID | Vv_12:6053035-6053039:- |
| Species | Vistis vinifera | Coordinates | 12:6053035..6053039 |
| Microexon Cluster ID | MEP09 |
| Size | 5 |
| Phase | 2 |
| Pfam Domain Motif | SKG6 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 19,34,5,50 |
| Microexon location in the Microexon-tag | 3 |
| Microexon-tag DNA Seq | GTDTWYATTCCDGSMMRAGATSAAAATGGAARYTATCSRCCCYTRMMAWCMAGCWCAGGAWTATCARGTGGAGYYATWGSTGGMATAKYTRTAGSAGYAGTAGYWGKR |
| Logo of Microexon-tag DNA Seq | 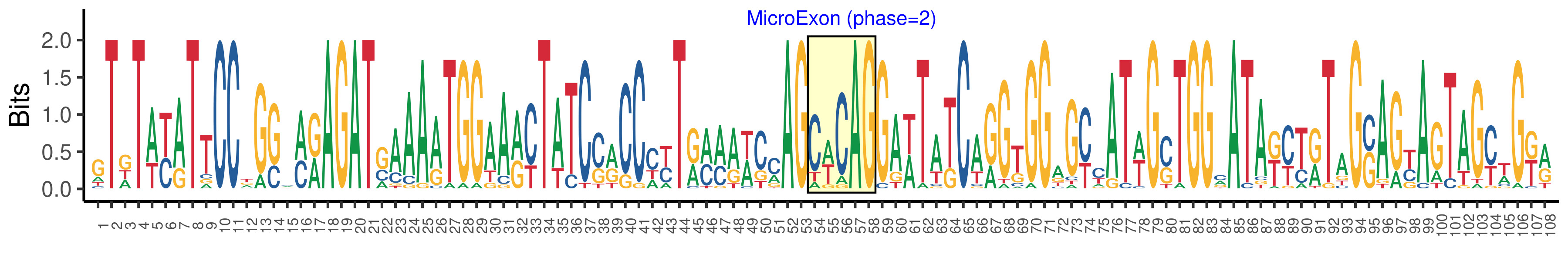 |
| Alignment of exons | 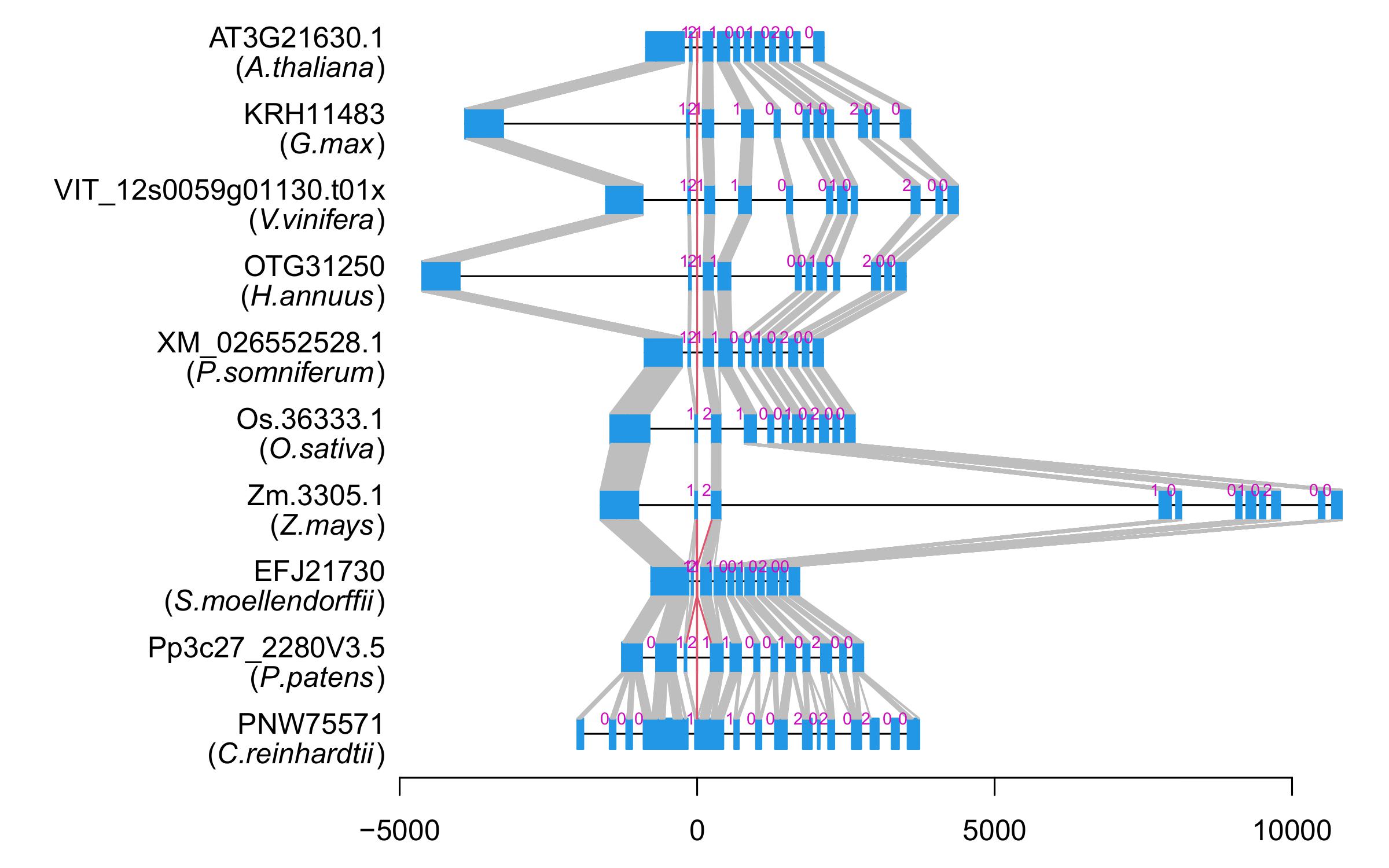 |
| Microexon DNA seq | CACAG |
| Microexon Amino Acid seq | STG |
| Microexon-tag DNA Seq | GTTTACATTCCTACTAAAGATACCAGTGGAAGCTATCGGGCCCTGAAATCCAGCACAGGATTAGCAGGTGGTGTGATTGCTGGCATATCTATAGCAGCAGTAGTAGGA |
| Microexon-tag Amino Acid Seq | VYIPTKDTSGSYRALKSSTGLAGGVIAGISIAAVVG |
| Microexon-tag spanning region | 6052855-6053972 |
| Microexon-tag prediction score | 0.9471 |
| Overlapped with the annotated transcript (%) | 76.6 |
| New Transcript ID | VIT_12s0059g01130.t01x |
| Reference Transcript ID | VIT_12s0059g01130.t01 |
| Gene ID | VIT_12s0059g01130 |
| Gene Name | NA |
Vv_12:6053035-6053039:- does not have available information here.
| Microexon DNA seq | CACAG |
| Microexon Amino Acid seq | STG |
| Microexon-tag DNA Seq | GTTTACATTCCTACTAAAGATACCAGTGGAAGCTATCGGGCCCTGAAATCCAGCACAGGATTAGCAGGTGGTGTGATTGCTGGCATATCTATAGCAGCAGTAGTAGGA |
| Microexon-tag Amino Acid seq | VYIPTKDTSGSYRALKSSTGLAGGVIAGISIAAVVG |
| Transcript ID | Vv.5163.2 |
| Gene ID | Vv.5163 |
| Gene Name | NA |
| Pfam domain motif | VSP |
| Motif E-value | 0.084 |
| Motif start | 194 |
| Motif end | 237 |
| Protein seq | >Vv.5163.2 MKQKVGLGFFVLLSVFCAVDSQCSRGCDLALGSYYVWQGSNLTFISQLFQTTISEILSYNSQIANQDSVEADTRIRVPYS SCDCINGEFLGKVFNYTVQSGDTYDLVAETYYSNLTTSAWLQNFNSYAANQIPDTDAYLNVTLNCSCGNSTVSKDYGLFL SYPLRPEDNLTSVAESEGLNASLLQSYNPDSNFSAGSGLVYIPTKDTSGSYRALKSSTGLAGGVIAGISIAAVVGVLLLT VCIYIGFYRKRKVKEAALLPTEEHSLQPGHGPGIASDKAVESTGPAFGSSAGLTGITVDKSVEFSYEELAKASDNFNLAN KIGQGGFGSVYYAELRGEKAAIKKMDMQASREFLAELKVLTHVHHLNLVRLIGYCVEGSLFLVYEYIENGNLSQHLRGSG RDPLQWSSRVQIALDSARGLEYIHEHTVPVYIHRDIKSANILIDKNFHGKVADFGLTKLTEVGSSSLPTRLVGTFGYMPP EYAQYGDVSPKVDVYAFGVVLYELISAKEAVVKDNGSVAESKGLVALFEDVLNKPDPREDLRKLVDPRLEDNYPLDSVRK MAQLAKACTQENPQLRPSMRTIVVALMTLSSSTEDWDVGSFYDNQALVNLMSGR* |
| CDS seq | >Vv.5163.2 ATGAAACAGAAGGTGGGTTTAGGGTTTTTTGTACTGCTCTCCGTTTTCTGTGCAGTTGATTCGCAGTGCAGTCGCGGCTG TGATCTTGCTCTGGGCTCATACTATGTCTGGCAAGGTTCCAACCTCACTTTTATCTCTCAGCTATTCCAGACAACGATTT CTGAAATTCTCAGCTACAACTCACAAATCGCTAATCAAGATAGTGTTGAAGCCGATACCAGAATCCGCGTGCCTTACTCC TCATGTGATTGCATCAATGGAGAATTTCTGGGCAAAGTGTTCAACTACACGGTGCAGTCCGGTGATACGTACGATCTTGT TGCGGAGACATACTATTCAAATCTGACCACTTCGGCGTGGTTGCAGAACTTCAATAGCTATGCCGCCAATCAAATTCCTG ATACGGATGCTTATCTGAATGTGACTTTAAATTGTTCTTGTGGCAATAGCACCGTTTCCAAGGATTATGGGTTGTTCTTG TCCTACCCGCTTCGCCCAGAGGATAATTTGACTTCGGTTGCAGAGTCAGAGGGTTTGAATGCTTCCTTGTTGCAGAGTTA CAATCCGGATTCCAATTTTAGTGCTGGGAGTGGGCTTGTTTACATTCCTACTAAAGATACCAGTGGAAGCTATCGGGCCC TGAAATCCAGCACAGGATTAGCAGGTGGTGTGATTGCTGGCATATCTATAGCAGCAGTAGTAGGAGTGCTGTTATTGACA GTTTGTATATATATTGGATTTTACAGAAAGAGGAAGGTGAAGGAAGCAGCATTGCTTCCAACTGAAGAACATTCTCTTCA ACCTGGGCATGGTCCTGGAATTGCTTCAGATAAAGCTGTAGAATCAACAGGTCCTGCTTTTGGTTCTTCTGCAGGTCTTA CAGGCATTACTGTGGACAAATCAGTGGAGTTCTCATATGAAGAACTTGCTAAGGCTTCTGATAATTTTAATTTGGCTAAT AAGATCGGTCAAGGTGGCTTTGGATCTGTTTACTATGCAGAGCTGAGGGGCGAGAAAGCTGCAATCAAGAAGATGGATAT GCAAGCATCAAGAGAATTTCTTGCTGAATTAAAAGTTTTGACACATGTGCATCACCTAAATTTGGTGCGCTTGATTGGAT ATTGTGTTGAGGGTTCTCTTTTTCTAGTCTATGAATACATTGAGAATGGCAACTTAAGCCAACATTTGCGTGGCTCAGGA AGGGACCCATTGCAATGGTCTAGTAGGGTTCAAATTGCCCTTGATTCAGCAAGAGGTCTTGAATATATTCATGAGCATAC TGTTCCTGTGTATATCCATCGTGATATTAAGTCAGCAAATATACTGATAGATAAGAATTTCCATGGAAAGGTTGCAGATT TTGGTCTAACAAAATTGACTGAAGTTGGGAGTTCATCCCTACCCACACGTCTTGTGGGTACATTTGGATACATGCCACCA GAATACGCTCAATATGGTGACGTCTCTCCCAAAGTAGATGTATATGCTTTTGGGGTTGTCCTTTATGAACTTATTTCTGC CAAGGAGGCTGTGGTCAAGGACAATGGTTCTGTTGCTGAATCAAAGGGCCTTGTGGCTTTGTTCGAGGATGTGCTAAATA AGCCAGATCCACGAGAGGACCTTCGCAAACTAGTTGACCCTAGGCTTGAAGATAACTACCCACTCGACTCTGTGCGCAAG ATGGCCCAACTTGCCAAAGCATGCACCCAGGAAAATCCTCAGCTAAGACCAAGCATGAGAACCATCGTGGTTGCCCTAAT GACTCTTTCATCCTCAACAGAGGACTGGGATGTTGGTTCCTTCTACGACAACCAAGCCCTCGTCAATCTAATGTCTGGAA GGTAG |