
| Microexon ID | Ps_NW_020622381.1:16853486-16853490:+ |
| Species | Papaver somniferum | Coordinates | NW_020622381.1:16853486..16853490 |
| Microexon Cluster ID | MEP08 |
| Size | 5 |
| Phase | 1 |
| Pfam Domain Motif | Peptidase_C1 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 52,5,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTYGAYGCWMGAACWGMTTGGYCTCADTGYASCACHATTGGRARMATWCTWGATCARGGWCAYTGTGGTTCTTGYTGGGCWTTTGGTGCTGTKGARKCACTRYCWGAT |
| Logo of Microexon-tag DNA Seq | 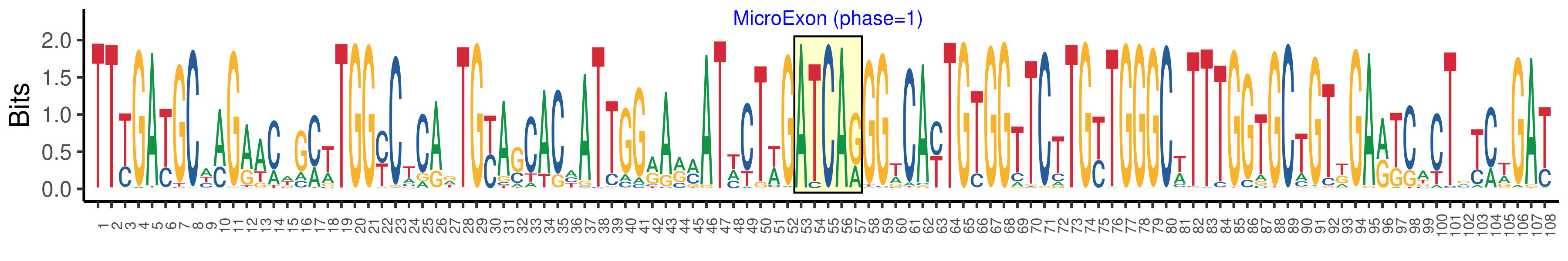 |
| Alignment of exons | 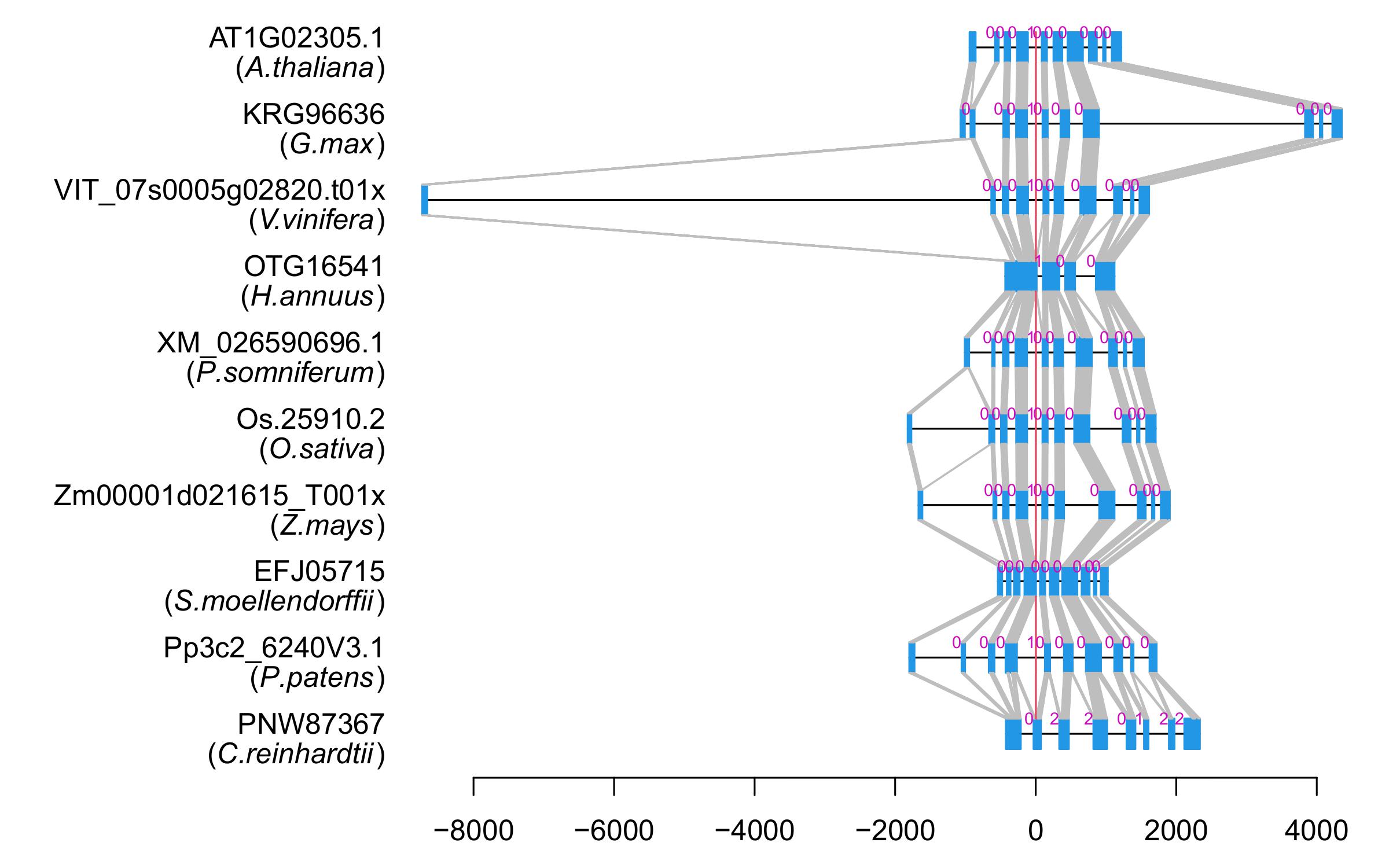 |
Ps_NW_020622381.1:16853486-16853490:+ does not have available information here.
| Transcript ID | XM_026579189.1 |
| Protein ID | XP_026434974.1 |
| Gene ID | LOC113332667 |
| Gene Name | NA |
| Pfam domain motif | Peptidase_C1 |
| Motif E-value | 1.5e-66 |
| Motif start | 95 |
| Motif end | 329 |
| Protein seq | >XP_026434974.1 MNMVPLVILILGTIFLAPNSQADATKSGPEILQDTIVQQINANPNAGWEAGLNSRFSNYTVSQFKYLLGVKPRTASSRND LLDVPIVSHPKNLELPKEFDARTAWPQCTTIGKILDQGHCGSCWAVAAAEALSDRFCVNLGMNIALSANDLLACCGFMCG DGCDGGYPISAWRYFVQNGIVTEACDPYFDAIGCSHPGCEPGFPTPKCVKRCKDKNQIWKQSKHFSVNAYQVGSDPTDIM AEVYKNGPVEVAFTVYEDFAHYKSGVYKHITGDVMGGHAVKLIGWGTTEDGEDFWLLANQWNRSWGDDGYFKIRRGTNEC GIEEEVVAGMPSTKNILIQKYAATADDFSYVPTDASI* |
| CDS seq | >XM_026579189.1 ATGAATATGGTGCCTCTGGTAATACTAATCTTGGGAACCATTTTCTTGGCTCCTAACTCACAGGCGGATGCAACAAAATC AGGCCCGGAGATTCTTCAGGACACCATTGTCCAACAGATAAATGCAAACCCTAATGCTGGATGGGAAGCTGGCTTGAATT CCCGCTTTTCAAATTACACAGTTTCCCAATTCAAGTACCTGTTGGGTGTAAAACCGAGAACAGCATCATCCCGCAATGAT TTGCTGGACGTTCCTATTGTCAGTCATCCCAAAAACTTGGAGTTGCCTAAGGAATTTGATGCCAGAACAGCTTGGCCTCA ATGTACTACCATCGGCAAAATTCTTGATCAGGGGCATTGTGGTTCTTGTTGGGCTGTTGCTGCAGCGGAAGCACTGTCTG ATCGTTTCTGCGTCAATCTTGGCATGAACATTGCCTTGTCTGCTAATGACCTCTTGGCATGTTGTGGCTTTATGTGCGGA GATGGTTGTGACGGTGGTTATCCTATTTCTGCGTGGCGATATTTTGTACAAAATGGCATTGTTACTGAAGCGTGTGATCC TTATTTTGATGCCATTGGATGTTCTCACCCTGGTTGTGAACCTGGATTTCCCACTCCTAAGTGTGTGAAACGATGCAAAG ATAAAAACCAGATCTGGAAGCAGTCAAAGCACTTCAGTGTCAATGCATACCAGGTCGGTTCCGACCCCACAGATATCATG GCAGAGGTCTACAAGAATGGTCCTGTCGAGGTGGCTTTCACTGTTTATGAGGATTTTGCTCATTACAAATCGGGCGTTTA CAAACACATCACAGGGGATGTGATGGGAGGACACGCTGTAAAGTTGATTGGTTGGGGAACTACTGAAGATGGAGAGGATT TTTGGCTTTTGGCCAACCAGTGGAATAGAAGCTGGGGAGACGACGGTTACTTCAAAATCAGGAGAGGTACAAATGAATGT GGAATTGAAGAGGAGGTGGTTGCTGGTATGCCTTCTACCAAGAACATTTTAATCCAGAAGTACGCTGCCACTGCTGATGA TTTTTCTTACGTTCCAACTGATGCCTCCATCTGA |
| Microexon DNA seq | ATCAG |
| Microexon Amino Acid seq | DQ |
| Microexon-tag DNA Seq | TTTGATGCCAGAACAGCTTGGCCTCAATGTACTACCATCGGCAAAATTCTTGATCAGGGGCATTGTGGTTCTTGTTGGGCTGTTGCTGCAGCGGAAGCACTGTCTGAT |
| Microexon-tag Amino Acid seq | FDARTAWPQCTTIGKILDQGHCGSCWAVAAAEALSD |
| Transcript ID | XM_026579189.1 |
| Gene ID | Ps.74101 |
| Gene Name | NA |
| Pfam domain motif | Peptidase_C1 |
| Motif E-value | 1.5e-66 |
| Motif start | 95 |
| Motif end | 329 |
| Protein seq | >XM_026579189.1 MNMVPLVILILGTIFLAPNSQADATKSGPEILQDTIVQQINANPNAGWEAGLNSRFSNYTVSQFKYLLGVKPRTASSRND LLDVPIVSHPKNLELPKEFDARTAWPQCTTIGKILDQGHCGSCWAVAAAEALSDRFCVNLGMNIALSANDLLACCGFMCG DGCDGGYPISAWRYFVQNGIVTEACDPYFDAIGCSHPGCEPGFPTPKCVKRCKDKNQIWKQSKHFSVNAYQVGSDPTDIM AEVYKNGPVEVAFTVYEDFAHYKSGVYKHITGDVMGGHAVKLIGWGTTEDGEDFWLLANQWNRSWGDDGYFKIRRGTNEC GIEEEVVAGMPSTKNILIQKYAATADDFSYVPTDASI* |
| CDS seq | >XM_026579189.1 ATGAATATGGTGCCTCTGGTAATACTAATCTTGGGAACCATTTTCTTGGCTCCTAACTCACAGGCGGATGCAACAAAATC AGGCCCGGAGATTCTTCAGGACACCATTGTCCAACAGATAAATGCAAACCCTAATGCTGGATGGGAAGCTGGCTTGAATT CCCGCTTTTCAAATTACACAGTTTCCCAATTCAAGTACCTGTTGGGTGTAAAACCGAGAACAGCATCATCCCGCAATGAT TTGCTGGACGTTCCTATTGTCAGTCATCCCAAAAACTTGGAGTTGCCTAAGGAATTTGATGCCAGAACAGCTTGGCCTCA ATGTACTACCATCGGCAAAATTCTTGATCAGGGGCATTGTGGTTCTTGTTGGGCTGTTGCTGCAGCGGAAGCACTGTCTG ATCGTTTCTGCGTCAATCTTGGCATGAACATTGCCTTGTCTGCTAATGACCTCTTGGCATGTTGTGGCTTTATGTGCGGA GATGGTTGTGACGGTGGTTATCCTATTTCTGCGTGGCGATATTTTGTACAAAATGGCATTGTTACTGAAGCGTGTGATCC TTATTTTGATGCCATTGGATGTTCTCACCCTGGTTGTGAACCTGGATTTCCCACTCCTAAGTGTGTGAAACGATGCAAAG ATAAAAACCAGATCTGGAAGCAGTCAAAGCACTTCAGTGTCAATGCATACCAGGTCGGTTCCGACCCCACAGATATCATG GCAGAGGTCTACAAGAATGGTCCTGTCGAGGTGGCTTTCACTGTTTATGAGGATTTTGCTCATTACAAATCGGGCGTTTA CAAACACATCACAGGGGATGTGATGGGAGGACACGCTGTAAAGTTGATTGGTTGGGGAACTACTGAAGATGGAGAGGATT TTTGGCTTTTGGCCAACCAGTGGAATAGAAGCTGGGGAGACGACGGTTACTTCAAAATCAGGAGAGGTACAAATGAATGT GGAATTGAAGAGGAGGTGGTTGCTGGTATGCCTTCTACCAAGAACATTTTAATCCAGAAGTACGCTGCCACTGCTGATGA TTTTTCTTACGTTCCAACTGATGCCTCCATCTGA |