
| Microexon ID | Ps_NC_039359.1:20830120-20830124:- |
| Species | Papaver somniferum | Coordinates | NC_039359.1:20830120..20830124 |
| Microexon Cluster ID | MEP08 |
| Size | 5 |
| Phase | 1 |
| Pfam Domain Motif | Peptidase_C1 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 52,5,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTYGAYGCWMGAACWGMTTGGYCTCADTGYASCACHATTGGRARMATWCTWGATCARGGWCAYTGTGGTTCTTGYTGGGCWTTTGGTGCTGTKGARKCACTRYCWGAT |
| Logo of Microexon-tag DNA Seq | 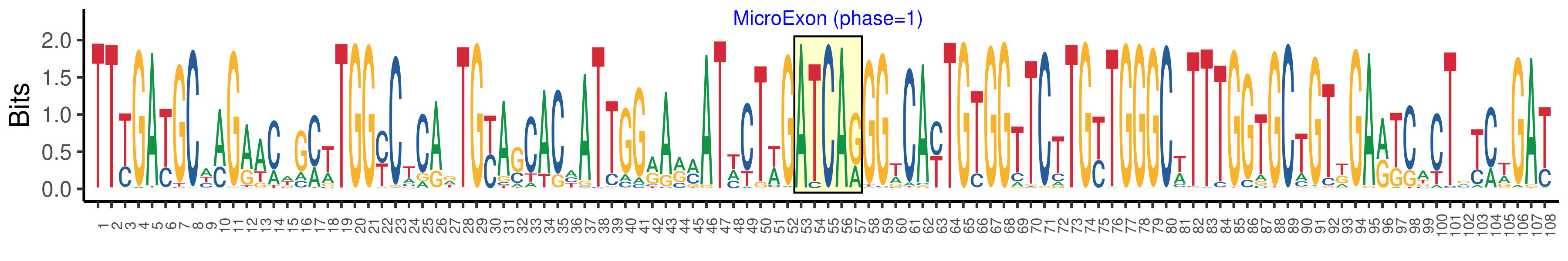 |
| Alignment of exons |  |
Ps_NC_039359.1:20830120-20830124:- does not have available information here.
| Transcript ID | XM_026590695.1 |
| Protein ID | XP_026446480.1 |
| Gene ID | LOC113347104 |
| Gene Name | NA |
| Pfam domain motif | Peptidase_C1 |
| Motif E-value | 1.3e-66 |
| Motif start | 95 |
| Motif end | 329 |
| Protein seq | >XP_026446480.1 MNMVPLVILILGTIFLAPNPQADAIKSGPEILQDTIVQQINANPNAGWEAGLNSRFSNYTVSQFKYLLGVKPRTASSRND LLDVPIVSHPKNLELPKEFDARTAWPQCTTIGKILDQGHCGSCWAVAAAEALSDRFCVNLGMNIALSANDLLACCGFMCG DGCDGGYPISAWRYFVQNGIVTEACDPYFDAIGCSHPGCEPGFPTPKCVKQCKDKNQIWKQSKHFSVNAYQVGSDPIDIM AEVYKNGPVEVAFTVYEDFAHYKSGVYKHITGDVMGGHAVKLIGWGTTEDGEDYWLLANQWNRSWGDDGYFKIRRGTNEC GIEEDVVAGMPSTKNILIQKYAATADDFSDVPTDASV* |
| CDS seq | >XM_026590695.1 ATGAATATGGTGCCTCTGGTAATCCTAATCCTGGGAACCATTTTCTTGGCTCCTAACCCACAGGCGGATGCAATAAAATC AGGCCCGGAGATTCTTCAGGACACCATTGTCCAACAGATAAATGCAAACCCTAATGCTGGATGGGAAGCTGGCTTGAATT CCCGCTTTTCAAATTACACAGTTTCCCAATTCAAGTACCTATTGGGTGTAAAACCGAGAACAGCATCATCCCGCAATGAT TTGCTGGACGTTCCTATTGTCAGTCACCCCAAAAACTTGGAGTTGCCTAAGGAATTTGATGCCAGAACAGCTTGGCCTCA ATGTACCACCATCGGCAAAATTCTTGATCAGGGGCATTGTGGTTCTTGTTGGGCTGTTGCTGCAGCGGAAGCACTGTCTG ATCGTTTCTGCGTCAATCTTGGCATGAACATTGCCTTGTCTGCTAATGACCTCTTGGCATGTTGTGGCTTTATGTGCGGA GATGGTTGTGACGGTGGTTATCCTATTTCTGCGTGGCGATATTTTGTACAAAATGGCATTGTTACTGAAGCGTGTGATCC TTACTTTGATGCCATTGGATGTTCTCACCCTGGTTGTGAACCTGGATTTCCCACTCCTAAGTGTGTGAAACAATGCAAAG ATAAAAACCAGATCTGGAAGCAGTCAAAGCACTTCAGTGTCAATGCATACCAGGTCGGTTCCGACCCCATAGATATCATG GCAGAGGTCTACAAGAATGGTCCTGTCGAGGTGGCTTTCACTGTTTATGAGGATTTTGCTCATTACAAATCGGGCGTTTA CAAACACATCACAGGGGATGTGATGGGAGGACACGCTGTAAAGTTGATTGGTTGGGGAACTACTGAAGATGGAGAGGATT ATTGGCTTTTGGCCAACCAGTGGAATAGAAGCTGGGGAGATGACGGTTACTTCAAAATCAGGAGAGGTACAAATGAATGT GGAATTGAAGAGGATGTGGTTGCTGGTATGCCTTCTACCAAGAACATTTTAATCCAGAAGTACGCTGCCACTGCTGATGA TTTTTCTGACGTTCCAACTGATGCCTCCGTCTGA |
| Microexon DNA seq | ATCAG |
| Microexon Amino Acid seq | DQ |
| Microexon-tag DNA Seq | TTTGATGCCAGAACAGCTTGGCCTCAATGTACCACCATCGGCAAAATTCTTGATCAGGGGCATTGTGGTTCTTGTTGGGCTGTTGCTGCTGCAGAAGCACTGTCTGAT |
| Microexon-tag Amino Acid seq | FDARTAWPQCTTIGKILDQGHCGSCWAVAAAEALSD |
| Transcript ID | Ps.8971.2 |
| Gene ID | Ps.8971 |
| Gene Name | NA |
| Pfam domain motif | Peptidase_C1 |
| Motif E-value | 2.4e-65 |
| Motif start | 95 |
| Motif end | 329 |
| Protein seq | >Ps.8971.2 MNMVPLVILILGTIFLAPNPQADAIKSGPEILQDTIVQQINANPNAGWEAGLNSRFSNYTVSQFKYLLGVKPRTASSRND LLDVPIVSHPKNLELPKEFDARTAWPQCTTIGKILDQGHCGSCWAVAAAEALSDRFCVNLGMNITLSANDLLACCGFMCG NGCGGGVPIFAWRYFVHHGIVTEECDPYFDAIGCSHPGCEPGFPTPKCVKHCKDKNQIWKQSKHFGVNAYHVKSNPTDIM AEVYKNGPVEVAFTVYEDFAHYKSGVYKHITGDVMGGHAVKLIGWGTTEDGEDYWLLANQWNRSWGDDGYFKIRRGTNEC GIERGVFAGMPSTKNILIRKYAADDDDSYVPNDASI* |
| CDS seq | >Ps.8971.2 ATGAATATGGTGCCTCTGGTAATCCTAATCCTGGGAACCATTTTCTTGGCTCCTAACCCACAGGCGGATGCAATAAAATC AGGCCCGGAGATTCTTCAGGACACCATTGTCCAACAGATAAATGCAAACCCTAATGCTGGATGGGAAGCTGGCTTGAATT CCCGCTTTTCAAATTACACAGTTTCCCAATTCAAGTACCTATTGGGTGTAAAACCGAGAACAGCATCATCCCGCAATGAT TTGCTGGACGTTCCTATTGTCAGTCACCCCAAAAACTTGGAGTTGCCTAAGGAATTTGATGCCAGAACAGCTTGGCCTCA ATGTACCACCATCGGCAAAATTCTTGATCAGGGGCATTGTGGTTCTTGTTGGGCTGTTGCTGCTGCAGAAGCACTGTCTG ATCGTTTCTGCGTCAATCTTGGCATGAACATTACCTTATCTGCTAATGACCTCTTGGCATGTTGTGGCTTTATGTGCGGA AATGGTTGTGGAGGTGGTGTTCCTATTTTTGCATGGCGATATTTTGTACACCATGGCATTGTTACTGAAGAGTGTGATCC TTACTTCGATGCCATTGGATGTTCTCACCCTGGTTGTGAACCTGGATTTCCCACTCCTAAGTGCGTGAAACATTGCAAAG ATAAAAACCAGATCTGGAAGCAGTCAAAGCACTTCGGTGTCAATGCATATCATGTCAAATCCAACCCCACTGATATCATG GCAGAGGTCTACAAAAATGGTCCTGTCGAGGTGGCTTTCACTGTTTATGAGGATTTTGCTCATTACAAATCGGGTGTTTA CAAGCACATCACAGGGGATGTAATGGGAGGTCATGCTGTAAAGTTGATTGGCTGGGGAACTACTGAAGACGGAGAGGATT ATTGGCTTTTGGCCAACCAGTGGAATAGAAGCTGGGGAGATGACGGTTACTTCAAAATCAGGAGAGGTACAAATGAATGT GGAATTGAAAGGGGTGTGTTTGCTGGTATGCCTTCTACCAAGAACATTTTAATCCGGAAATATGCTGCTGATGATGATGA TTCTTACGTTCCAAATGATGCTTCCATCTGA |