
| Microexon ID | Pp_2:4098935-4098939:- |
| Species | Physcomitrium patens | Coordinates | 2:4098935..4098939 |
| Microexon Cluster ID | MEP08 |
| Size | 5 |
| Phase | 1 |
| Pfam Domain Motif | Peptidase_C1 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 52,5,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTYGAYGCWMGAACWGMTTGGYCTCADTGYASCACHATTGGRARMATWCTWGATCARGGWCAYTGTGGTTCTTGYTGGGCWTTTGGTGCTGTKGARKCACTRYCWGAT |
| Logo of Microexon-tag DNA Seq | 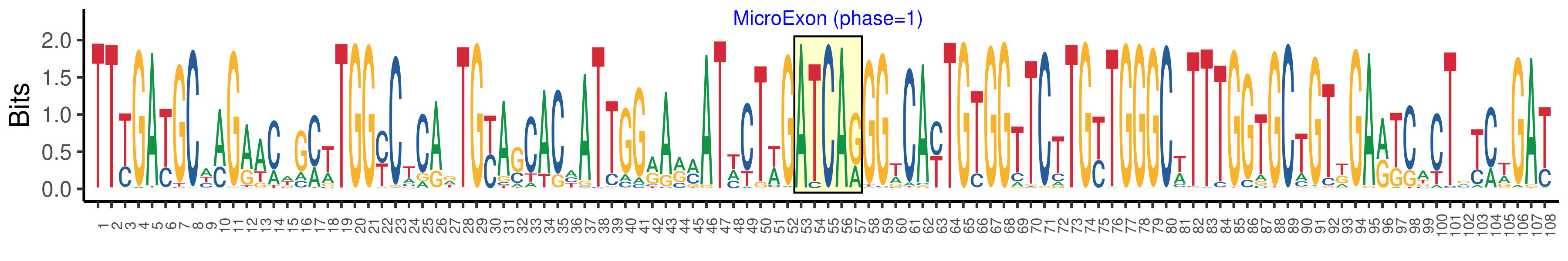 |
| Alignment of exons | 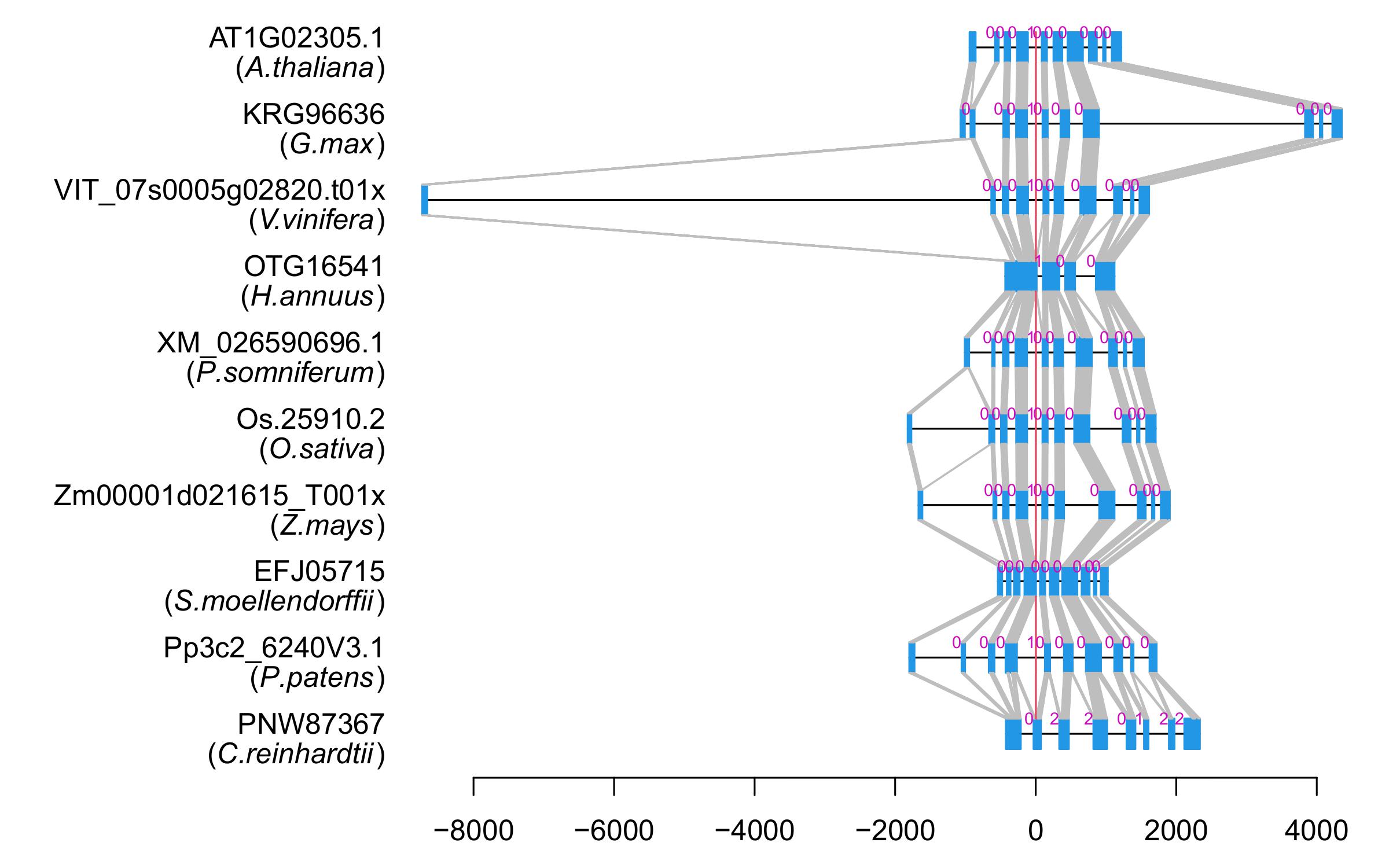 |
| Microexon DNA seq | ATCAG |
| Microexon Amino Acid seq | DQ |
| Microexon-tag DNA Seq | TTTGATGCCCGTAAACAATGGAGTCATTGTCCAACAATTGGTGACATTCTAGATCAGGGTCACTGCGGATCATGCTGGGCTTTTGGGGCTGTTGAGTCACTGACTGAT |
| Microexon-tag Amino Acid Seq | FDARKQWSHCPTIGDILDQGHCGSCWAFGAVESLTD |
| Microexon-tag spanning region | 4098758-4099257 |
| Microexon-tag prediction score | 0.9309 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Pp3c2_6240V3.1x |
| Reference Transcript ID | Pp3c2_6240V3.1 |
| Gene ID | Pp3c2_6240 |
| Gene Name | NA |
| Transcript ID | Pp3c2_6240V3.1 |
| Protein ID | Pp3c2_6240V3.1 |
| Gene ID | Pp3c2_6240 |
| Gene Name | NA |
| Pfam domain motif | Peptidase_C1 |
| Motif E-value | 4.8e-67 |
| Motif start | 103 |
| Motif end | 336 |
| Protein seq | >Pp3c2_6240V3.1 MAYERMGKLDLSLLLMLCALFFAVQAGRLEPELLGNNRLIHQQALVDKVNAHPGATWTAGFNERFAKHTIEHLKKMCGAI LTPANKLEPSIETISHKHKKLYLPKEFDARKQWSHCPTIGDILDQGHCGSCWAFGAVESLTDRFCIHLNESVSLSENDLL ACCGFECGYGCEGGYPIRAWKYFKHSGVVTNKCDPYFDQKGCAHPGCYPTYETPKCEKQCVDDEFWVQSKHLGVNAYEMS MEPEDLMAELYTNGPVEVAFEVYEDFAHYKTGVYKHLFGGFMGGHAVKLIGWGTTDDGVDYWTIVNSWNTNWGEDGLFRI VRGNDECGIESNAVAGLPSRKGLHSAM* |
| CDS seq | >Pp3c2_6240V3.1 ATGGCGTATGAGAGGATGGGGAAGCTGGATTTGAGCTTGCTGTTGATGCTTTGCGCTTTGTTCTTCGCAGTTCAGGCAGG TAGGCTTGAGCCTGAGCTACTTGGAAACAACAGACTCATCCATCAGCAAGCGCTTGTGGACAAAGTCAATGCCCATCCAG GAGCGACCTGGACGGCGGGCTTCAATGAGCGATTCGCAAAGCACACAATTGAGCACTTGAAGAAGATGTGCGGTGCTATC TTGACACCCGCTAACAAGTTGGAGCCTTCTATCGAAACGATCAGTCACAAGCACAAGAAGTTATATCTACCCAAAGAGTT TGATGCCCGTAAACAATGGAGTCATTGTCCAACAATTGGTGACATTCTAGATCAGGGTCACTGCGGATCATGCTGGGCTT TTGGGGCTGTTGAGTCACTGACTGATCGTTTCTGCATTCACTTGAATGAAAGTGTGTCACTATCAGAGAACGATCTCTTA GCATGCTGTGGATTCGAGTGCGGCTATGGCTGCGAGGGTGGGTACCCTATAAGAGCCTGGAAATACTTCAAGCACTCCGG AGTTGTGACAAACAAGTGTGATCCATACTTTGATCAGAAAGGCTGTGCTCACCCAGGCTGCTATCCTACCTATGAGACAC CTAAGTGCGAAAAGCAGTGTGTAGACGATGAGTTTTGGGTGCAGTCGAAGCATCTTGGTGTGAATGCGTACGAGATGTCC ATGGAACCTGAGGACCTCATGGCTGAGCTCTATACCAATGGTCCTGTTGAGGTGGCCTTTGAGGTTTACGAGGACTTCGC TCATTACAAAACTGGAGTTTACAAACATCTTTTCGGTGGCTTCATGGGAGGGCACGCTGTGAAGCTTATTGGATGGGGAA CCACCGACGATGGTGTGGACTACTGGACCATAGTGAACTCCTGGAACACGAACTGGGGAGAGGATGGTCTCTTCCGTATT GTGAGAGGCAACGATGAGTGTGGCATTGAGAGCAATGCAGTAGCTGGTCTCCCATCCAGAAAAGGTCTTCATTCTGCCAT GTGA |
| Microexon DNA seq | ATCAG |
| Microexon Amino Acid seq | DQ |
| Microexon-tag DNA Seq | TTTGATGCCCGTAAACAATGGAGTCATTGTCCAACAATTGGTGACATTCTAGATCAGGGTCACTGCGGATCATGCTGGGCTTTTGGGGCTGTTGAGTCACTGACTGAT |
| Microexon-tag Amino Acid seq | FDARKQWSHCPTIGDILDQGHCGSCWAFGAVESLTD |
| Transcript ID | Pp3c2_6240V3.1 |
| Gene ID | Pp.11006 |
| Gene Name | NA |
| Pfam domain motif | Peptidase_C1 |
| Motif E-value | 4.8e-67 |
| Motif start | 103 |
| Motif end | 336 |
| Protein seq | >Pp3c2_6240V3.1 MAYERMGKLDLSLLLMLCALFFAVQAGRLEPELLGNNRLIHQQALVDKVNAHPGATWTAGFNERFAKHTIEHLKKMCGAI LTPANKLEPSIETISHKHKKLYLPKEFDARKQWSHCPTIGDILDQGHCGSCWAFGAVESLTDRFCIHLNESVSLSENDLL ACCGFECGYGCEGGYPIRAWKYFKHSGVVTNKCDPYFDQKGCAHPGCYPTYETPKCEKQCVDDEFWVQSKHLGVNAYEMS MEPEDLMAELYTNGPVEVAFEVYEDFAHYKTGVYKHLFGGFMGGHAVKLIGWGTTDDGVDYWTIVNSWNTNWGEDGLFRI VRGNDECGIESNAVAGLPSRKGLHSAM* |
| CDS seq | >Pp3c2_6240V3.1 ATGGCGTATGAGAGGATGGGGAAGCTGGATTTGAGCTTGCTGTTGATGCTTTGCGCTTTGTTCTTCGCAGTTCAGGCAGG TAGGCTTGAGCCTGAGCTACTTGGAAACAACAGACTCATCCATCAGCAAGCGCTTGTGGACAAAGTCAATGCCCATCCAG GAGCGACCTGGACGGCGGGCTTCAATGAGCGATTCGCAAAGCACACAATTGAGCACTTGAAGAAGATGTGCGGTGCTATC TTGACACCCGCTAACAAGTTGGAGCCTTCTATCGAAACGATCAGTCACAAGCACAAGAAGTTATATCTACCCAAAGAGTT TGATGCCCGTAAACAATGGAGTCATTGTCCAACAATTGGTGACATTCTAGATCAGGGTCACTGCGGATCATGCTGGGCTT TTGGGGCTGTTGAGTCACTGACTGATCGTTTCTGCATTCACTTGAATGAAAGTGTGTCACTATCAGAGAACGATCTCTTA GCATGCTGTGGATTCGAGTGCGGCTATGGCTGCGAGGGTGGGTACCCTATAAGAGCCTGGAAATACTTCAAGCACTCCGG AGTTGTGACAAACAAGTGTGATCCATACTTTGATCAGAAAGGCTGTGCTCACCCAGGCTGCTATCCTACCTATGAGACAC CTAAGTGCGAAAAGCAGTGTGTAGACGATGAGTTTTGGGTGCAGTCGAAGCATCTTGGTGTGAATGCGTACGAGATGTCC ATGGAACCTGAGGACCTCATGGCTGAGCTCTATACCAATGGTCCTGTTGAGGTGGCCTTTGAGGTTTACGAGGACTTCGC TCATTACAAAACTGGAGTTTACAAACATCTTTTCGGTGGCTTCATGGGAGGGCACGCTGTGAAGCTTATTGGATGGGGAA CCACCGACGATGGTGTGGACTACTGGACCATAGTGAACTCCTGGAACACGAACTGGGGAGAGGATGGTCTCTTCCGTATT GTGAGAGGCAACGATGAGTGTGGCATTGAGAGCAATGCAGTAGCTGGTCTCCCATCCAGAAAAGGTCTTCATTCTGCCAT GTGA |