
| Microexon ID | Gm_3:42855765-42855769:- |
| Species | Glycine max | Coordinates | 3:42855765..42855769 |
| Microexon Cluster ID | MEP08 |
| Size | 5 |
| Phase | 1 |
| Pfam Domain Motif | Peptidase_C1 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 52,5,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTYGAYGCWMGAACWGMTTGGYCTCADTGYASCACHATTGGRARMATWCTWGATCARGGWCAYTGTGGTTCTTGYTGGGCWTTTGGTGCTGTKGARKCACTRYCWGAT |
| Logo of Microexon-tag DNA Seq | 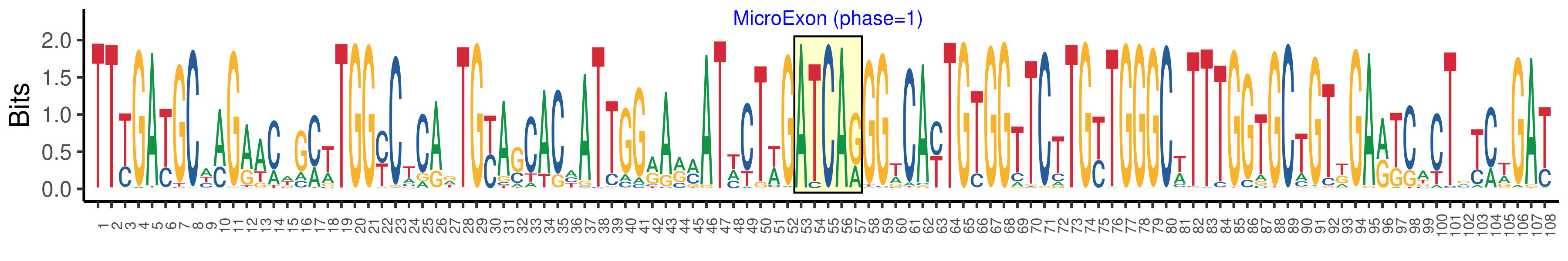 |
| Alignment of exons | 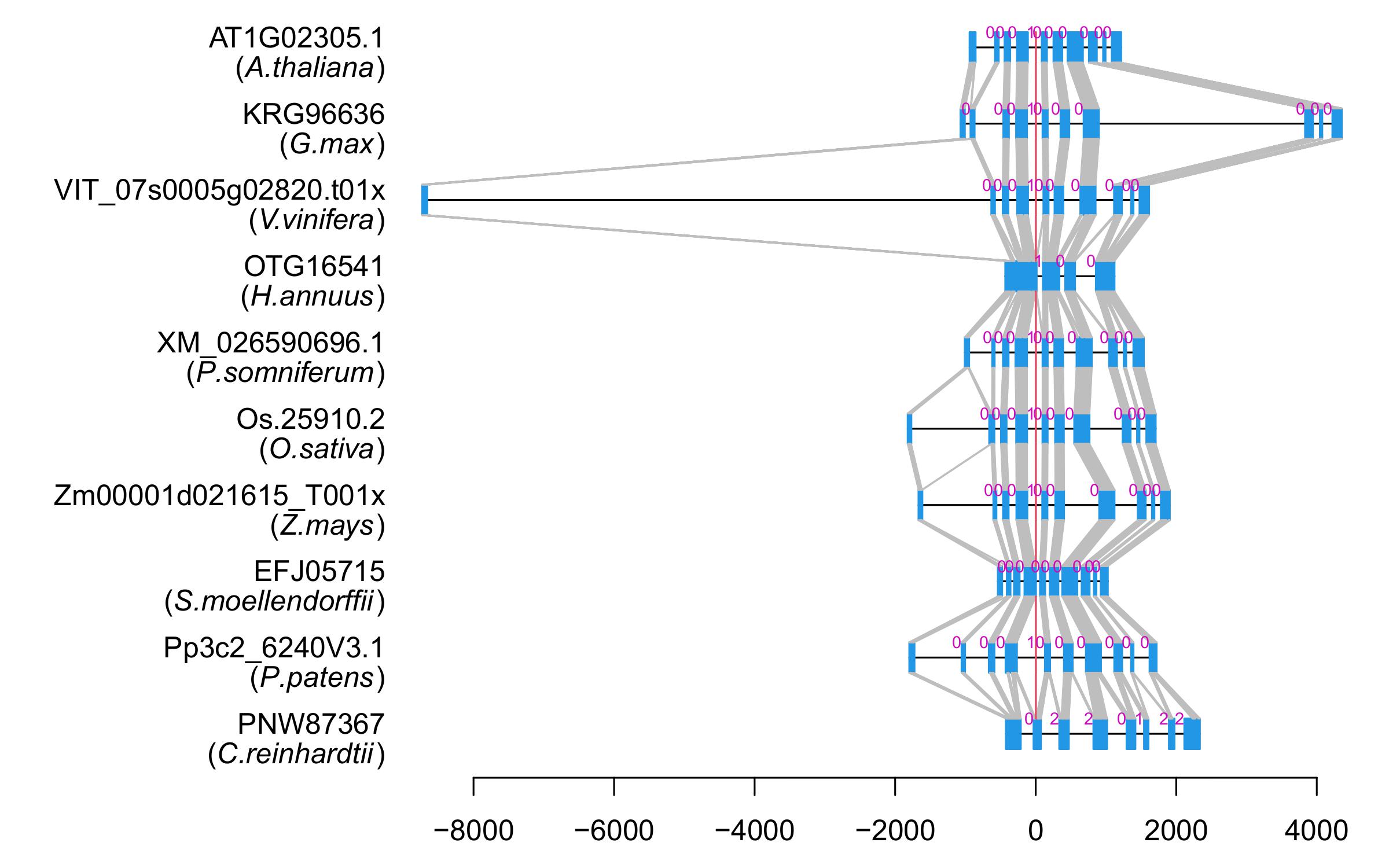 |
| Microexon DNA seq | ATCAG |
| Microexon Amino Acid seq | DQ |
| Microexon-tag DNA Seq | TTTGATGCGAGGACAGCTTGGTCGCAGTGTAGCACTATTGGAAGAATTCTAGATCAGGGTCACTGTGGTTCTTGTTGGGCATTTGGTGCTGTTGAATCATTGTCAGAT |
| Microexon-tag Amino Acid Seq | FDARTAWSQCSTIGRILDQGHCGSCWAFGAVESLSD |
| Microexon-tag spanning region | 42855620-42855949 |
| Microexon-tag prediction score | 0.9833 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH68369x |
| Reference Transcript ID | KRH68369 |
| Gene ID | GLYMA_03G226300 |
| Gene Name | NA |
| Transcript ID | KRH68369 |
| Protein ID | KRH68369 |
| Gene ID | GLYMA_03G226300 |
| Gene Name | NA |
| Pfam domain motif | Peptidase_C1 |
| Motif E-value | 5.4e-66 |
| Motif start | 101 |
| Motif end | 334 |
| Protein seq | >KRH68369 MASTHLLPLATFFLLLSASYLQIAGAEAQPLTSLKLNSHILQESTAKEINENPEAGWEAAINPRFSNYTVEQFKRLLGVK PMPKKELRSTPAISHPKTLKLPKNFDARTAWSQCSTIGRILDQGHCGSCWAFGAVESLSDRFCIHFDVNISLSVNDLLAC CGFLCGSGCDGGYPLYAWRYLAHHGVVTEECDPYFDQIGCSHPGCEPAYRTPKCVKKCVSGNQVWKKSKHYSVSAYRVNS DPHDIMAEVYKNGPVEVAFTVYEDFAYYKSGVYKHITGYELGGHAVKLIGWGTTDDGEDYWLLANQWNREWGDDGYFKIR RGTNECGIEEDVTAGLPSTKNLVREVTDMDADAAVSF* |
| CDS seq | >KRH68369 ATGGCTTCAACTCATCTGCTTCCCCTAGCAACCTTCTTCTTACTTCTCTCCGCTTCTTATCTCCAGATTGCTGGGGCGGA AGCACAACCGTTGACTAGTCTCAAGCTTAACTCTCATATTCTTCAGGAGTCTACTGCTAAAGAGATTAATGAAAATCCCG AGGCAGGATGGGAGGCTGCTATAAATCCTCGTTTCTCCAATTATACAGTTGAACAATTTAAGCGCCTTCTTGGAGTCAAA CCAATGCCTAAGAAGGAACTTAGAAGTACACCTGCTATATCTCACCCAAAGACGTTGAAATTGCCAAAGAATTTTGATGC GAGGACAGCTTGGTCGCAGTGTAGCACTATTGGAAGAATTCTAGATCAGGGTCACTGTGGTTCTTGTTGGGCATTTGGTG CTGTTGAATCATTGTCAGATCGCTTTTGCATTCATTTTGATGTTAATATCTCTCTCTCTGTTAATGACCTTCTTGCATGC TGTGGCTTTCTGTGTGGATCTGGCTGTGATGGAGGATATCCCTTATATGCATGGCGATACTTAGCCCACCATGGTGTTGT CACTGAAGAGTGTGACCCATATTTTGATCAAATTGGCTGTTCTCATCCTGGTTGTGAGCCAGCTTATCGGACTCCCAAGT GTGTTAAAAAGTGTGTAAGTGGAAACCAAGTTTGGAAGAAGTCAAAGCACTATAGTGTTAGTGCATACAGAGTCAACTCT GATCCCCATGATATCATGGCGGAAGTTTACAAGAATGGGCCAGTTGAAGTTGCATTTACCGTTTATGAGGATTTTGCTTA CTACAAATCAGGAGTTTACAAACACATCACTGGTTATGAACTAGGTGGTCATGCAGTAAAGCTAATTGGGTGGGGAACAA CTGATGATGGGGAGGACTATTGGCTTCTCGCAAATCAGTGGAACAGAGAATGGGGAGATGATGGTTACTTCAAGATCCGA AGAGGGACAAACGAATGTGGGATTGAAGAGGATGTAACAGCTGGTTTGCCTTCCACCAAAAACCTCGTCAGAGAGGTGAC TGATATGGATGCTGACGCTGCTGTTTCATTCTGA |
| Microexon DNA seq | ATCAG |
| Microexon Amino Acid seq | DQ |
| Microexon-tag DNA Seq | TTTGATGCGAGGACAGCTTGGTCGCAGTGTAGCACTATTGGAAGAATTCTAGATCAGGGTCACTGTGGTTCTTGTTGGGCATTTGGTGCTGTTGAATCATTGTCAGAT |
| Microexon-tag Amino Acid seq | FDARTAWSQCSTIGRILDQGHCGSCWAFGAVESLSD |
| Transcript ID | KRH68369 |
| Gene ID | Gm.36953 |
| Gene Name | NA |
| Pfam domain motif | Peptidase_C1 |
| Motif E-value | 5.4e-66 |
| Motif start | 101 |
| Motif end | 334 |
| Protein seq | >KRH68369 MASTHLLPLATFFLLLSASYLQIAGAEAQPLTSLKLNSHILQESTAKEINENPEAGWEAAINPRFSNYTVEQFKRLLGVK PMPKKELRSTPAISHPKTLKLPKNFDARTAWSQCSTIGRILDQGHCGSCWAFGAVESLSDRFCIHFDVNISLSVNDLLAC CGFLCGSGCDGGYPLYAWRYLAHHGVVTEECDPYFDQIGCSHPGCEPAYRTPKCVKKCVSGNQVWKKSKHYSVSAYRVNS DPHDIMAEVYKNGPVEVAFTVYEDFAYYKSGVYKHITGYELGGHAVKLIGWGTTDDGEDYWLLANQWNREWGDDGYFKIR RGTNECGIEEDVTAGLPSTKNLVREVTDMDADAAVSF* |
| CDS seq | >KRH68369 ATGGCTTCAACTCATCTGCTTCCCCTAGCAACCTTCTTCTTACTTCTCTCCGCTTCTTATCTCCAGATTGCTGGGGCGGA AGCACAACCGTTGACTAGTCTCAAGCTTAACTCTCATATTCTTCAGGAGTCTACTGCTAAAGAGATTAATGAAAATCCCG AGGCAGGATGGGAGGCTGCTATAAATCCTCGTTTCTCCAATTATACAGTTGAACAATTTAAGCGCCTTCTTGGAGTCAAA CCAATGCCTAAGAAGGAACTTAGAAGTACACCTGCTATATCTCACCCAAAGACGTTGAAATTGCCAAAGAATTTTGATGC GAGGACAGCTTGGTCGCAGTGTAGCACTATTGGAAGAATTCTAGATCAGGGTCACTGTGGTTCTTGTTGGGCATTTGGTG CTGTTGAATCATTGTCAGATCGCTTTTGCATTCATTTTGATGTTAATATCTCTCTCTCTGTTAATGACCTTCTTGCATGC TGTGGCTTTCTGTGTGGATCTGGCTGTGATGGAGGATATCCCTTATATGCATGGCGATACTTAGCCCACCATGGTGTTGT CACTGAAGAGTGTGACCCATATTTTGATCAAATTGGCTGTTCTCATCCTGGTTGTGAGCCAGCTTATCGGACTCCCAAGT GTGTTAAAAAGTGTGTAAGTGGAAACCAAGTTTGGAAGAAGTCAAAGCACTATAGTGTTAGTGCATACAGAGTCAACTCT GATCCCCATGATATCATGGCGGAAGTTTACAAGAATGGGCCAGTTGAAGTTGCATTTACCGTTTATGAGGATTTTGCTTA CTACAAATCAGGAGTTTACAAACACATCACTGGTTATGAACTAGGTGGTCATGCAGTAAAGCTAATTGGGTGGGGAACAA CTGATGATGGGGAGGACTATTGGCTTCTCGCAAATCAGTGGAACAGAGAATGGGGAGATGATGGTTACTTCAAGATCCGA AGAGGGACAAACGAATGTGGGATTGAAGAGGATGTAACAGCTGGTTTGCCTTCCACCAAAAACCTCGTCAGAGAGGTGAC TGATATGGATGCTGACGCTGCTGTTTCATTCTGA |