
| Microexon ID | At_4:695701-695705:+ |
| Species | Arabidopsis thaliana | Coordinates | 4:695701..695705 |
| Microexon Cluster ID | MEP08 |
| Size | 5 |
| Phase | 1 |
| Pfam Domain Motif | Peptidase_C1 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 52,5,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTYGAYGCWMGAACWGMTTGGYCTCADTGYASCACHATTGGRARMATWCTWGATCARGGWCAYTGTGGTTCTTGYTGGGCWTTTGGTGCTGTKGARKCACTRYCWGAT |
| Logo of Microexon-tag DNA Seq | 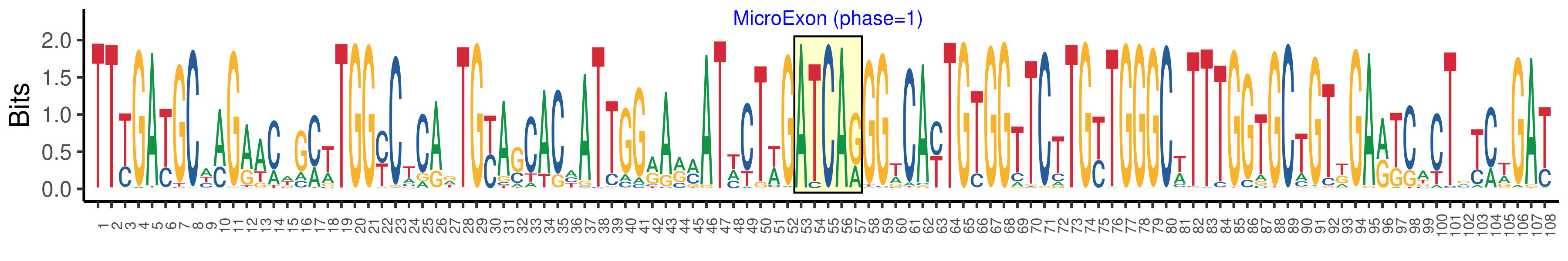 |
| Alignment of exons | 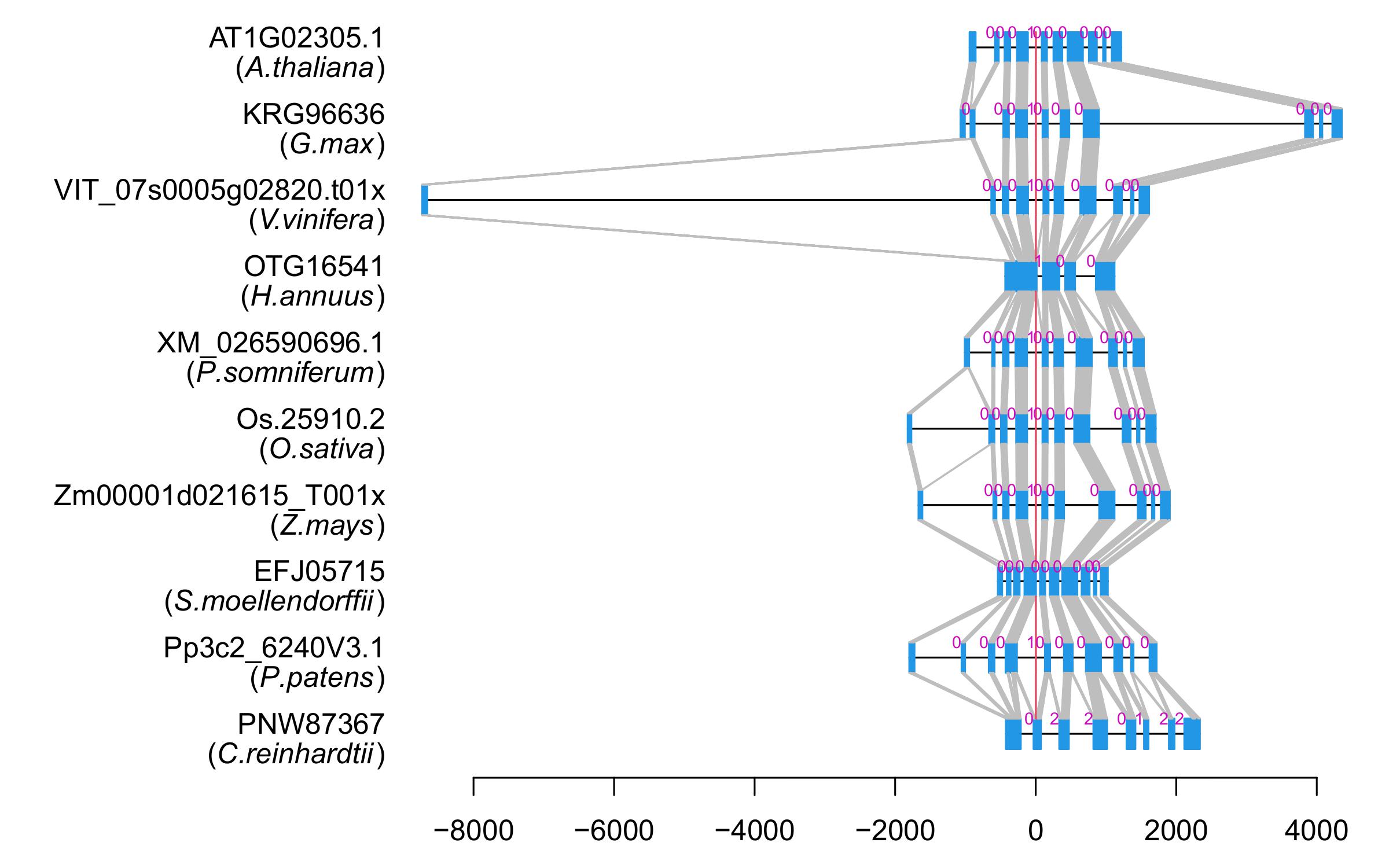 |
| Microexon DNA seq | ATCAG |
| Microexon Amino Acid seq | DQ |
| Microexon-tag DNA Seq | TTTGATGCTAGAACTGCTTGGCCACAGTGCACTAGTATTGGAAACATCTTAGATCAGGGACATTGTGGTTCTTGCTGGGCATTTGGTGCTGTTGAATCACTATCAGAT |
| Microexon-tag Amino Acid Seq | FDARTAWPQCTSIGNILDQGHCGSCWAFGAVESLSD |
| Microexon-tag spanning region | 695543-695845 |
| Microexon-tag prediction score | 0.9769 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | AT4G01610.1x |
| Reference Transcript ID | AT4G01610.1 |
| Gene ID | AT4G01610 |
| Gene Name | CATHB3 |
| Transcript ID | AT4G01610.1 |
| Protein ID | AT4G01610.1 |
| Gene ID | AT4G01610 |
| Gene Name | CATHB3 |
| Pfam domain motif | Peptidase_C1 |
| Motif E-value | 8.2e-67 |
| Motif start | 103 |
| Motif end | 337 |
| Protein seq | >AT4G01610.1 MAVYNTKLCLASVFLLLGLLLAFDLKGIEAESLTKQKLDSKILQDEIVKKVNENPNAGWKAAINDRFSNATVAEFKRLLG VKPTPKKHFLGVPIVSHDPSLKLPKAFDARTAWPQCTSIGNILDQGHCGSCWAFGAVESLSDRFCIQFGMNISLSVNDLL ACCGFRCGDGCDGGYPIAAWQYFSYSGVVTEECDPYFDNTGCSHPGCEPAYPTPKCSRKCVSDNKLWSESKHYSVSTYTV KSNPQDIMAEVYKNGPVEVSFTVYEDFAHYKSGVYKHITGSNIGGHAVKLIGWGTSSEGEDYWLMANQWNRGWGDDGYFM IRRGTNECGIEDEPVAGLPSSKNVFRVDTGSNDLPVASV* |
| CDS seq | >AT4G01610.1 ATGGCTGTTTACAATACCAAACTCTGTTTGGCCTCTGTTTTCTTGCTCTTAGGGCTTCTCTTGGCTTTTGACTTGAAGGG TATAGAAGCAGAAAGTCTTACCAAACAGAAACTGGACTCGAAGATACTTCAGGATGAGATTGTGAAGAAAGTCAACGAAA ACCCAAACGCTGGATGGAAAGCTGCTATAAATGATCGATTTTCAAACGCCACTGTTGCAGAGTTTAAGCGTCTTCTCGGT GTTAAACCAACACCAAAGAAGCATTTCTTAGGTGTTCCTATTGTGAGCCATGATCCATCTTTGAAGCTACCTAAAGCTTT TGATGCTAGAACTGCTTGGCCACAGTGCACTAGTATTGGAAACATCTTAGATCAGGGACATTGTGGTTCTTGCTGGGCAT TTGGTGCTGTTGAATCACTATCAGATAGATTCTGTATCCAATTTGGCATGAACATTTCTTTATCAGTAAATGATCTCCTA GCTTGTTGTGGATTCCGTTGTGGTGATGGTTGTGACGGTGGCTATCCAATTGCTGCTTGGCAATACTTTTCGTATAGCGG TGTTGTCACAGAAGAGTGTGATCCATACTTTGATAATACCGGATGCTCTCACCCGGGTTGCGAACCGGCATATCCTACAC CGAAATGTTCGAGGAAATGCGTTAGCGACAACAAACTATGGAGCGAGTCGAAGCATTACAGTGTTAGTACATACACTGTG AAATCTAATCCACAAGATATCATGGCAGAGGTTTATAAGAATGGACCTGTTGAAGTCTCTTTTACCGTTTATGAGGATTT TGCTCATTACAAATCTGGAGTCTATAAGCACATAACAGGTTCTAACATTGGAGGTCATGCTGTTAAACTTATTGGTTGGG GAACTAGCAGTGAAGGAGAAGATTATTGGTTGATGGCAAATCAATGGAACAGAGGCTGGGGTGATGATGGTTATTTCATG ATAAGGAGAGGAACTAATGAATGTGGAATTGAAGATGAACCAGTAGCTGGTTTGCCTTCAAGCAAGAATGTGTTTAGGGT TGATACCGGTTCGAATGATCTTCCGGTTGCATCGGTTTAA |
| Microexon DNA seq | ATCAG |
| Microexon Amino Acid seq | DQ |
| Microexon-tag DNA Seq | TTTGATGCTAGAACTGCTTGGCCACAGTGCACTAGTATTGGAAACATCTTAGATCAGGGACATTGTGGTTCTTGCTGGGCATTTGGTGCTGTTGAATCACTATCAGAT |
| Microexon-tag Amino Acid seq | FDARTAWPQCTSIGNILDQGHCGSCWAFGAVESLSD |
| Transcript ID | AT4G01610.1 |
| Gene ID | At.17637 |
| Gene Name | CATHB3 |
| Pfam domain motif | Peptidase_C1 |
| Motif E-value | 8.3e-67 |
| Motif start | 103 |
| Motif end | 337 |
| Protein seq | >AT4G01610.1 MAVYNTKLCLASVFLLLGLLLAFDLKGIEAESLTKQKLDSKILQDEIVKKVNENPNAGWKAAINDRFSNATVAEFKRLLG VKPTPKKHFLGVPIVSHDPSLKLPKAFDARTAWPQCTSIGNILDQGHCGSCWAFGAVESLSDRFCIQFGMNISLSVNDLL ACCGFRCGDGCDGGYPIAAWQYFSYSGVVTEECDPYFDNTGCSHPGCEPAYPTPKCSRKCVSDNKLWSESKHYSVSTYTV KSNPQDIMAEVYKNGPVEVSFTVYEDFAHYKSGVYKHITGSNIGGHAVKLIGWGTSSEGEDYWLMANQWNRGWGDDGYFM IRRGTNECGIEDEPVAGLPSSKNVFRVDTGSNDLPVASV* |
| CDS seq | >AT4G01610.1 ATGGCTGTTTACAATACCAAACTCTGTTTGGCCTCTGTTTTCTTGCTCTTAGGGCTTCTCTTGGCTTTTGACTTGAAGGG TATAGAAGCAGAAAGTCTTACCAAACAGAAACTGGACTCGAAGATACTTCAGGATGAGATTGTGAAGAAAGTCAACGAAA ACCCAAACGCTGGATGGAAAGCTGCTATAAATGATCGATTTTCAAACGCCACTGTTGCAGAGTTTAAGCGTCTTCTCGGT GTTAAACCAACACCAAAGAAGCATTTCTTAGGTGTTCCTATTGTGAGCCATGATCCATCTTTGAAGCTACCTAAAGCTTT TGATGCTAGAACTGCTTGGCCACAGTGCACTAGTATTGGAAACATCTTAGATCAGGGACATTGTGGTTCTTGCTGGGCAT TTGGTGCTGTTGAATCACTATCAGATAGATTCTGTATCCAATTTGGCATGAACATTTCTTTATCAGTAAATGATCTCCTA GCTTGTTGTGGATTCCGTTGTGGTGATGGTTGTGACGGTGGCTATCCAATTGCTGCTTGGCAATACTTTTCGTATAGCGG TGTTGTCACAGAAGAGTGTGATCCATACTTTGATAATACCGGATGCTCTCACCCGGGTTGCGAACCGGCATATCCTACAC CGAAATGTTCGAGGAAATGCGTTAGCGACAACAAACTATGGAGCGAGTCGAAGCATTACAGTGTTAGTACATACACTGTG AAATCTAATCCACAAGATATCATGGCAGAGGTTTATAAGAATGGACCTGTTGAAGTCTCTTTTACCGTTTATGAGGATTT TGCTCATTACAAATCTGGAGTCTATAAGCACATAACAGGTTCTAACATTGGAGGTCATGCTGTTAAACTTATTGGTTGGG GAACTAGCAGTGAAGGAGAAGATTATTGGTTGATGGCAAATCAATGGAACAGAGGCTGGGGTGATGATGGTTATTTCATG ATAAGGAGAGGAACTAATGAATGTGGAATTGAAGATGAACCAGTAGCTGGTTTGCCTTCAAGCAAGAATGTGTTTAGGGT TGATACCGGTTCGAATGATCTTCCGGTTGCATCGGTTTAA |