
| Microexon ID | Ha_6:53799251-53799255:- |
| Species | Helianthus annuus | Coordinates | 6:53799251..53799255 |
| Microexon Cluster ID | MEP06 |
| Size | 5 |
| Phase | 0 |
| Pfam Domain Motif | Ribul_P_3_epim |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 51,5,52 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCYSAGCGCATGSTCMRYTKYGGYGCYGAYTGGCTYCACATGGACATCATGGATGGGCAYTTTGTYCCWAAYYTWACWATTGGTGCTCCAGTYATYGAGAGTTTGAGR |
| Logo of Microexon-tag DNA Seq | 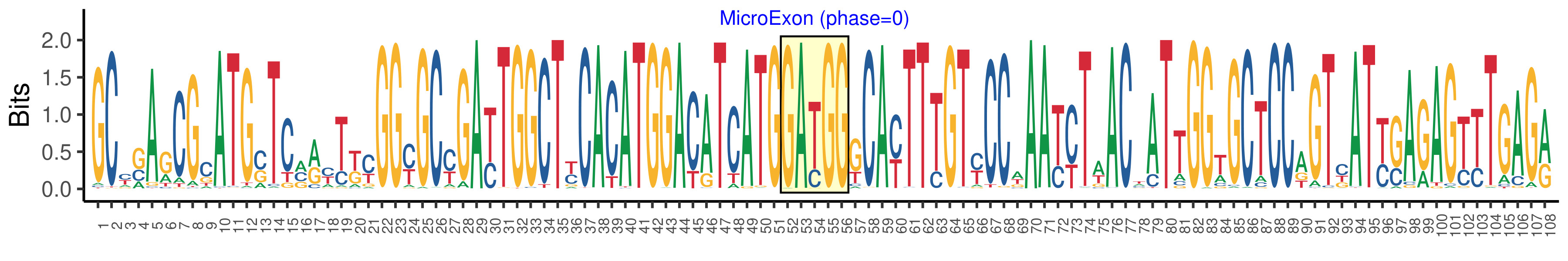 |
| Alignment of exons | 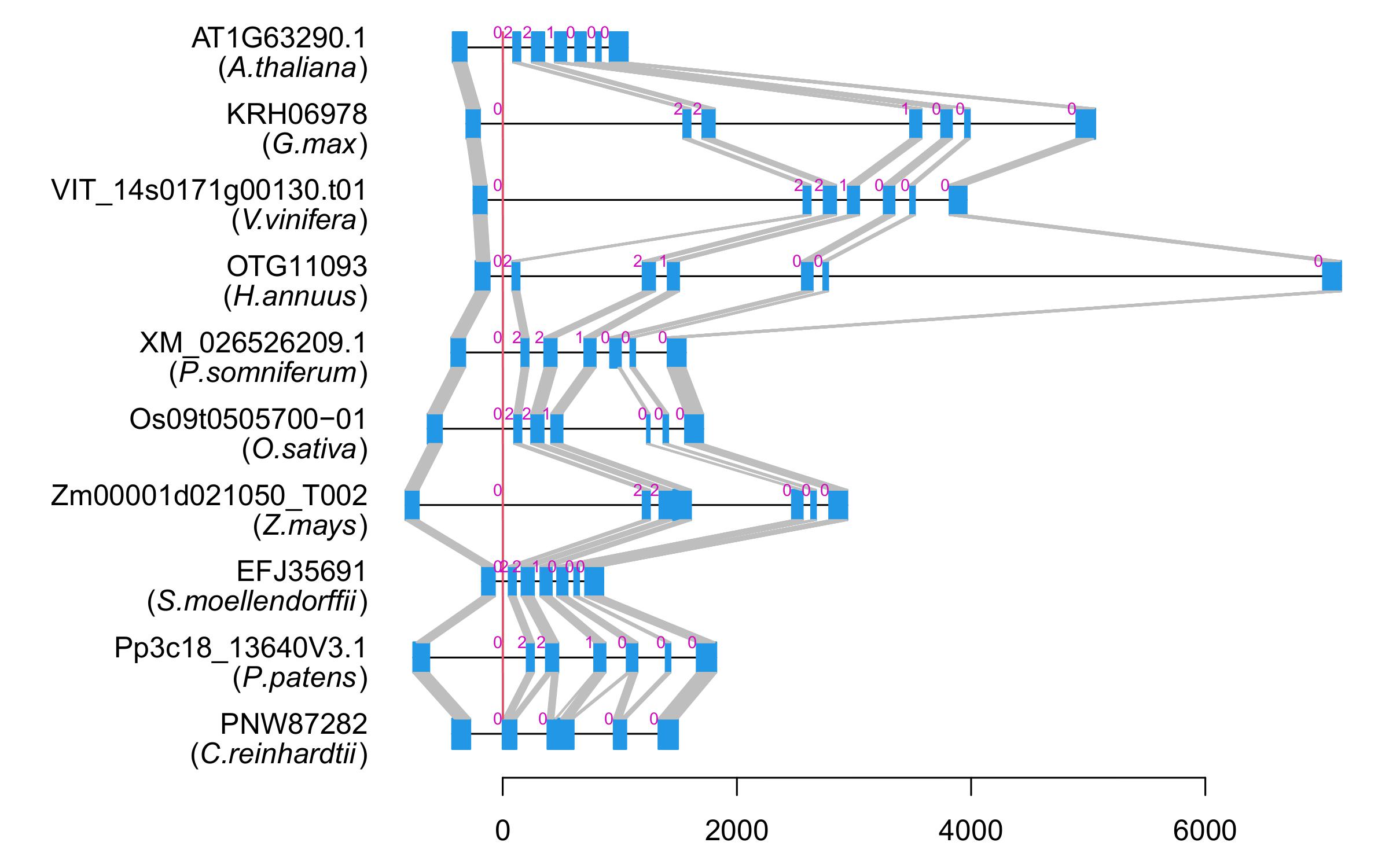 |
| Microexon DNA seq | GATGG |
| Microexon Amino Acid seq | DG |
| Microexon-tag DNA Seq | GCTGAGCGAATGGTCAACTTCGGTGCTGACTGGCTTCACATGGACATCATGGATGGCCATATTGTCCCAAATCTAACTATTGGTGCTCCAGTTATTGAGAGTTTGAGA |
| Microexon-tag Amino Acid Seq | AERMVNFGADWLHMDIMDGHIVPNLTIGAPVIESLR |
| Microexon-tag spanning region | 53799121-53799404 |
| Microexon-tag prediction score | 0.9732 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | OTG23145x |
| Reference Transcript ID | OTG23145 |
| Gene ID | HannXRQ_Chr06g0179181 |
| Gene Name | NA |
| Transcript ID | OTG23145 |
| Protein ID | OTG23145 |
| Gene ID | HannXRQ_Chr06g0179181 |
| Gene Name | NA |
| Pfam domain motif | Ribul_P_3_epim |
| Motif E-value | 5.2e-15 |
| Motif start | 1 |
| Motif end | 53 |
| Protein seq | >OTG23145 MLSSDFANIAFEAERMVNFGADWLHMDIMDGHIVPNLTIGAPVIESLRKHTKECMGRFCLKTITVTHMLVNG* |
| CDS seq | >OTG23145 ATGCTATCATCAGACTTCGCTAATATTGCATTCGAGGCTGAGCGAATGGTCAACTTCGGTGCTGACTGGCTTCACATGGA CATCATGGATGGCCATATTGTCCCAAATCTAACTATTGGTGCTCCAGTTATTGAGAGTTTGAGAAAGCACACAAAGGAGT GCATGGGTCGGTTTTGCCTCAAAACCATAACCGTAACCCATATGTTGGTTAATGGATAA |
| Microexon DNA seq | GATGG |
| Microexon Amino Acid seq | DG |
| Microexon-tag DNA Seq | GCTGAGCGAATGGTCAACTTCGGTGCTGACTGGCTTCACATGGACATCATGGATGGCCATATTGTCCCAAATCTAACTATTGGTGCTCCAGTTATTGAGAGTTTGAGA |
| Microexon-tag Amino Acid seq | AERMVNFGADWLHMDIMDGHIVPNLTIGAPVIESLR |
| Transcript ID | Ha.48496.4 |
| Gene ID | Ha.48496 |
| Gene Name | NA |
| Pfam domain motif | Ribul_P_3_epim |
| Motif E-value | 5.4e-15 |
| Motif start | 1 |
| Motif end | 53 |
| Protein seq | >Ha.48496.4 MLSSDFANIAFEAERMVNFGADWLHMDIMDGHIVPNLTIGAPVIESLRKHTKECMGRFCLKTITVTHMLVNG* |
| CDS seq | >Ha.48496.4 ATGCTATCATCAGACTTCGCTAATATTGCATTCGAGGCTGAGCGAATGGTCAACTTCGGTGCTGACTGGCTTCACATGGA CATCATGGATGGCCATATTGTCCCAAATCTAACTATTGGTGCTCCAGTTATTGAGAGTTTGAGAAAGCACACAAAGGAGT GCATGGGTCGGTTTTGCCTCAAAACCATAACCGTAACCCATATGTTGGTTAATGGATAA |