
| Microexon ID | Bn_scaffoldA08:21333325-21333328:- |
| Species | Brassica Napus | Coordinates | scaffoldA08:21333325..21333328 |
| Microexon Cluster ID | MEP05 |
| Size | 4 |
| Phase | 2 |
| Pfam Domain Motif | Helicase_C |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 53,4,51 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GATGAYTGGMTTTCWKCSARRAYWCAAGTTGTWGTKGCHACWGTRGCWTTTGGRATGGGWATWGATARRMARGATGTYMGDATTGTKTGYCAYTTYAAYWTKCCWAAR |
| Logo of Microexon-tag DNA Seq | 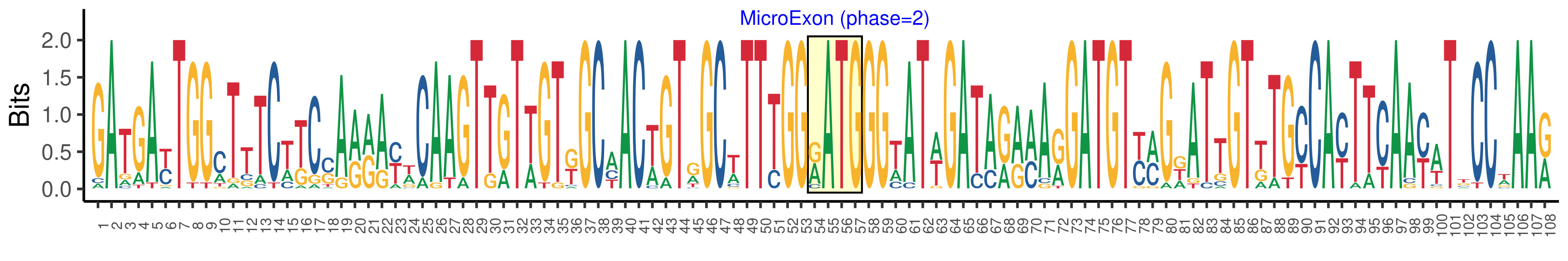 |
| Alignment of exons | 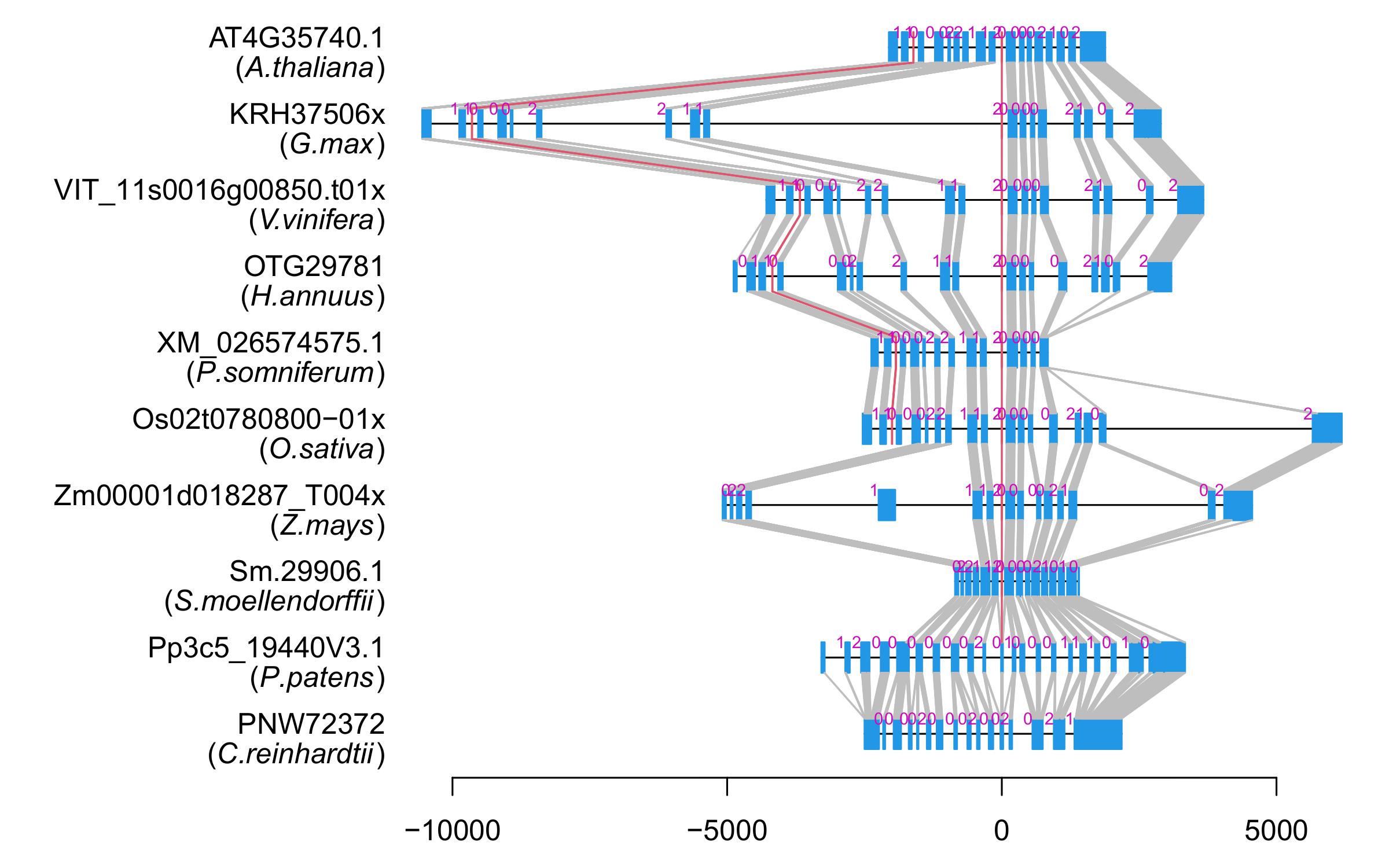 |
| Microexon DNA seq | AATG |
| Microexon Amino Acid seq | GM |
| Microexon-tag DNA Seq | GATGACTGGCTGTCCTCGAAGAAGCAAGTTATTGTTGCCACAGTGGCTTTCGGAATGGGTATAGATAAGAAGGATGTGAGAATGGTTTGCCACTTTAACGTCCCAAAA |
| Microexon-tag Amino Acid Seq | DDWLSSKKQVIVATVAFGMGIDKKDVRMVCHFNVPK |
| Microexon-tag spanning region | 21333187-21333509 |
| Microexon-tag prediction score | 0.9527 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | BnaA08T0178400ZSx |
| Reference Transcript ID | BnaA08T0178400ZS |
| Gene ID | BnaA08G0178400ZS |
| Gene Name | NA |
| Transcript ID | BnaA08T0178400ZS |
| Protein ID | |
| Gene ID | BnaA08G0178400ZS |
| Gene Name | |
| Pfam domain motif | Helicase_C |
| Motif E-value | 1.00E-16 |
| Motif start | 246 |
| Motif end | 350 |
| Protein seq | >BnaA08T0178400ZS MKKSPLPVQNVNRSDKNVAGKEALVKLLRWHFGHGDFRGKQLEAIQAVVSGRDCFCLMPTGGGKSICYQIPALAKPGIVL VVSPLIALMENQVMALKEKGIAAEYLSSTQATHVRNKIHEDLDSGKPSVRLLYVTPELIATKGFMLKLRKLHDRGLLNLI AIDEAHCISSWGHDFRPSYRQLSTLRDSLTDVPVLALTATAAPKVQKDVIDSLSLRNPLVLKSSFNRPNIFYEVRYKDLI DNAYTDLCNMLKSCGKICAIIYCLERTTCDDLSLHLTSTGISSAAYHAGLNSHLRSTVLDDWLSSKKQVIVATVAFGMGI DKKDVRMVCHFNVPKSMESFYQESGRAGRDQLPSRSVLYYGVDDRKKMEFLLRNSENKKSPSSSSKKPTSDFEQIVRYCE GSGCRRKKILESFGEEFPVQQCKKTCDACKHPNQVARSLEELTTTASRRHNSSRVFITSSSDNKTNEGQYSEFWNRNEDG SNSDEEISDSDDGADVVKSLAGPKLSRKLGVDEKLVLLERAEEKYNESNKQVKKSEKNAISETLRESSKQRLLNELTKVL QLLGVKEIDSQNASEFLESECYRKYSKAGKSFYYSQIASTVRWLGTASRDELMTRLSSMAREEEPSGEPILVTEPSQNIE QEDGTTYTAEPHVDETTQQLVISPSCSPIRLPEIPSFSEFVNRRKMKHSTEGSDGKKPSKIMKLK* |
| CDS seq | >BnaA08T0178400ZS ATGAAGAAATCGCCGTTGCCGGTGCAGAACGTTAACCGCTCCGATAAGAACGTCGCCGGAAAAGAAGCACTGGTGAAGCT ATTGCGATGGCACTTCGGCCATGGAGATTTCAGAGGGAAGCAGCTAGAGGCGATTCAAGCGGTTGTATCAGGAAGGGATT GCTTTTGCTTGATGCCGACGGGAGGAGGGAAATCGATTTGTTATCAGATACCTGCGTTGGCTAAACCTGGAATCGTGCTT GTTGTTTCTCCTTTGATTGCTTTGATGGAGAACCAAGTGATGGCGTTGAAGGAGAAAGGTATTGCTGCTGAGTATCTCTC GTCCACTCAAGCTACACATGTTAGGAACAAGATCCACGAGGACCTTGATTCTGGAAAACCTTCTGTAAGATTGCTGTATG TAACTCCAGAACTGATCGCGACAAAAGGATTTATGCTGAAGCTGAGAAAGCTCCATGACAGGGGTTTACTGAATCTTATA GCCATAGACGAGGCACATTGCATCTCATCATGGGGCCATGACTTCAGACCTAGCTATCGTCAACTTTCAACCTTAAGAGA TTCTTTGACTGATGTTCCAGTGTTGGCTCTAACCGCTACTGCTGCTCCTAAGGTACAAAAGGATGTCATTGACTCCTTGA GTTTGCGGAATCCTTTGGTCCTCAAGTCCTCTTTCAATCGCCCAAATATCTTCTATGAAGTTAGGTACAAAGACCTTATA GACAATGCTTATACTGATTTGTGCAACATGCTCAAGTCATGTGGAAAAATATGTGCAATTATCTATTGCCTTGAGCGTAC AACCTGTGATGACTTGTCTCTTCATCTCACCAGTACTGGGATCTCTTCTGCTGCTTATCATGCAGGACTCAATAGTCATC TGAGAAGTACTGTTTTAGATGACTGGCTGTCCTCGAAGAAGCAAGTTATTGTTGCCACAGTGGCTTTCGGAATGGGTATA GATAAGAAGGATGTGAGAATGGTTTGCCACTTTAACGTCCCAAAATCGATGGAATCTTTCTACCAAGAGTCCGGTAGAGC AGGTCGGGATCAGCTACCTTCAAGAAGTGTATTATACTATGGCGTAGACGATAGAAAGAAAATGGAGTTTCTACTACGGA ACTCGGAGAATAAGAAATCCCCATCCTCATCCTCAAAGAAGCCTACATCTGACTTCGAGCAGATTGTGAGATACTGCGAA GGTTCTGGATGCCGAAGGAAAAAGATTCTTGAGAGCTTTGGTGAAGAGTTTCCTGTACAGCAATGCAAGAAAACGTGTGA TGCATGTAAGCATCCAAATCAAGTTGCTCGTAGCTTGGAGGAGCTCACGACTACCGCCTCCCGAAGACACAACTCTTCTC GAGTTTTCATCACCAGCTCGAGTGATAATAAGACCAATGAAGGGCAGTACTCTGAGTTCTGGAATCGTAATGAAGATGGA AGCAATTCCGACGAGGAAATATCAGATTCAGATGATGGAGCTGATGTTGTCAAAAGCCTAGCAGGACCTAAACTATCGAG GAAGCTGGGAGTAGATGAGAAACTGGTTTTACTGGAGCGGGCTGAGGAAAAGTACAACGAAAGCAATAAACAGGTCAAAA AGTCAGAGAAGAATGCGATATCTGAAACACTAAGGGAATCAAGCAAGCAGAGACTCTTGAATGAGCTTACAAAAGTACTT CAACTGCTTGGTGTAAAAGAGATTGATTCCCAAAACGCATCTGAGTTTCTTGAAAGCGAGTGCTATAGAAAATACAGCAA AGCAGGGAAATCGTTTTACTACTCACAGATAGCGAGTACTGTGAGATGGTTAGGAACCGCAAGCAGAGACGAGCTAATGA CCCGACTTAGCTCAATGGCCAGAGAAGAAGAGCCATCAGGAGAACCAATCCTGGTGACTGAACCATCACAGAACATAGAG CAAGAAGATGGAACCACATACACAGCCGAGCCACATGTCGACGAAACAACACAGCAGCTAGTGATTAGCCCGAGCTGTTC GCCTATAAGGCTCCCAGAGATTCCGTCATTCTCAGAGTTTGTGAACCGTAGAAAGATGAAGCACTCTACCGAAGGTTCTG ATGGCAAGAAACCGTCAAAGATAATGAAGCTAAAGTAA |
Bn_scaffoldA08:21333325-21333328:- does not have available information here.