
| Microexon ID | Zm_3:170273227-170273230:+ |
| Species | Zea mays | Coordinates | 3:170273227..170273230 |
| Microexon Cluster ID | MEP04 |
| Size | 4 |
| Phase | 0 |
| Pfam Domain Motif | Ribonuclease_T2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 51,4,53 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RGYWMWYCATCAACHTGCYWYRGTGGAAARGGAYYATTYTGGGCTCATGAGTGGGARAARCATGGRACWTGYTCYTCYCCWGTWRTTCRWGATGAATAYARYTAYTTY |
| Logo of Microexon-tag DNA Seq | 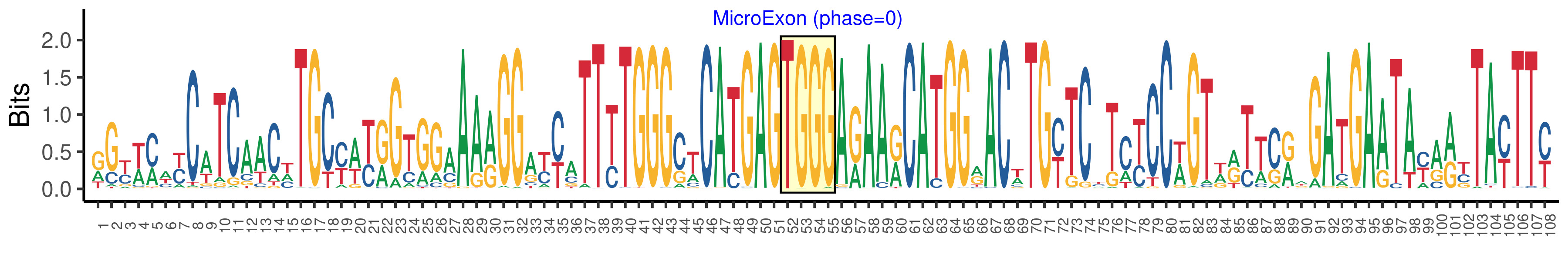 |
| Alignment of exons | 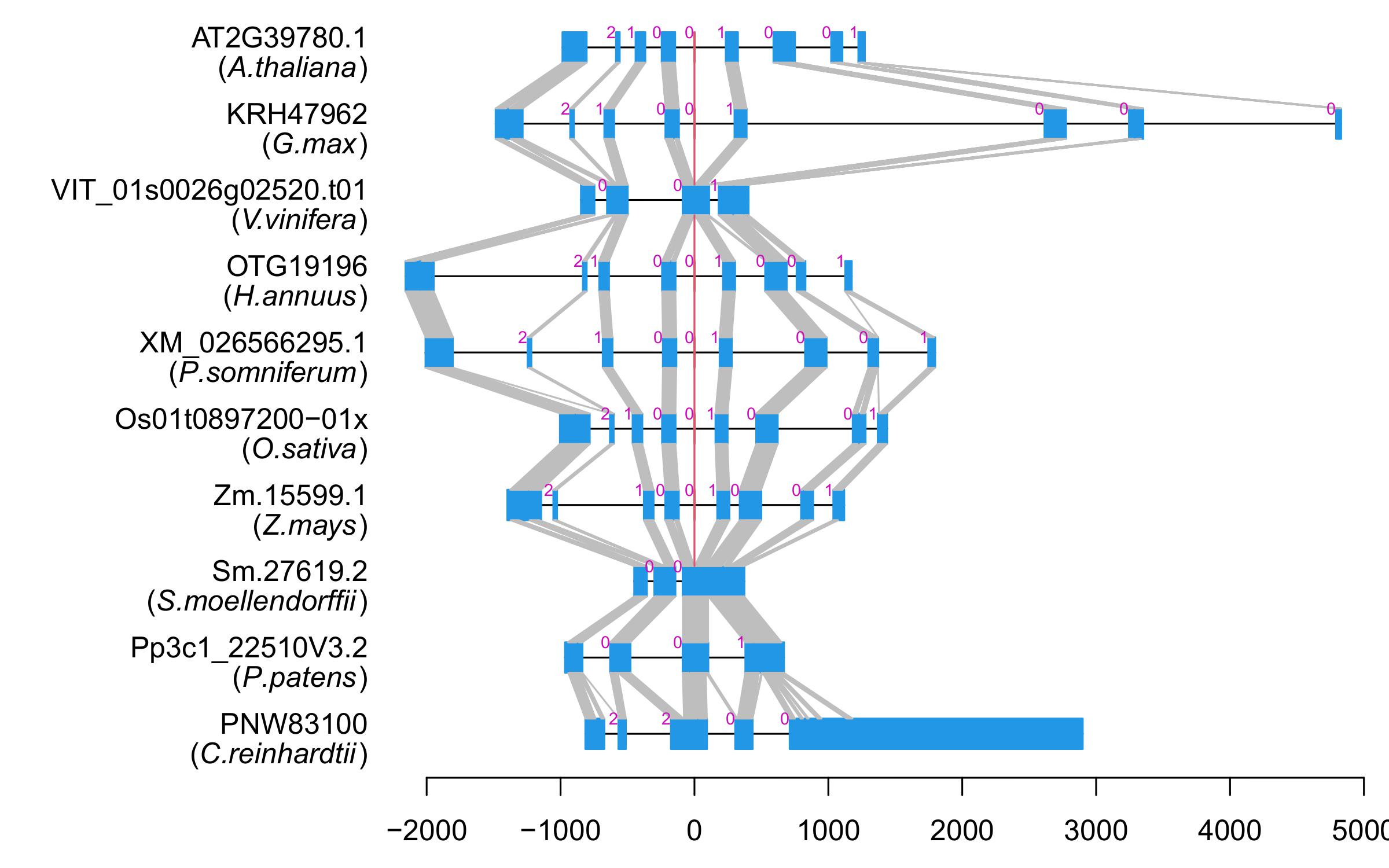 |
Zm_3:170273227-170273230:+ does not have available information here.
Zm_3:170273227-170273230:+ does not have available information here.
| Microexon DNA seq | TGGG |
| Microexon Amino Acid seq | WE |
| Microexon-tag DNA Seq | TCCAAATCTGGAACATGCTTCAGCGGCAAGGGACTCTTCTGGGCTCACGAGTGGGAGAAACATGGAACCTGCTCTGCCCCTGTGGTTCAGGATGAATTACAGTATTTC |
| Microexon-tag Amino Acid seq | SKSGTCFSGKGLFWAHEWEKHGTCSAPVVQDELQYF |
| Transcript ID | Zm.15600.1 |
| Gene ID | Zm.15600 |
| Gene Name | NA |
| Pfam domain motif | Ribonuclease_T2 |
| Motif E-value | 6.1e-46 |
| Motif start | 39 |
| Motif end | 225 |
| Protein seq | >Zm.15600.1 MTMATKAACVLAVWVLVASGLSDLGSARAPLGSKPQREFDYFALSLQWPGTICASTRHCCATNGCCRSEPLQTFTIHGLW PDYDDGTWPSCCRRTQFEMDKILPLKEVLDKYWPSLYCSKSGTCFSGKGLFWAHEWEKHGTCSAPVVQDELQYFTLALDL YFKYNVTEMLSSGWIQVSNGKEYALSDVIDTIKHAFGGSPQIVCKRGSIEELRLCFDKELKPRDCLTTSLANGSVSKSKH CPRYITLPTYDPLVLANSTVEIMTQFDEFEVPAALYTA* |
| CDS seq | >Zm.15600.1 ATGACGATGGCGACGAAGGCGGCGTGCGTGCTGGCGGTGTGGGTGCTCGTCGCATCGGGCCTGTCCGACCTGGGCTCAGC GCGCGCGCCCCTGGGCAGCAAGCCGCAGCGGGAGTTCGACTACTTCGCGCTCTCCCTCCAGTGGCCCGGCACCATCTGCG CCTCCACCCGCCACTGCTGCGCCACCAACGGCTGCTGCCGCTCGGAGCCTCTCCAGACGTTCACGATCCACGGGCTATGG CCGGACTACGACGACGGGACCTGGCCGTCGTGCTGCCGCCGCACCCAATTCGAGATGGACAAGATATTGCCACTGAAGGA GGTGCTTGACAAGTACTGGCCGTCCCTCTACTGCTCCAAATCTGGAACATGCTTCAGCGGCAAGGGACTCTTCTGGGCTC ACGAGTGGGAGAAACATGGAACCTGCTCTGCCCCTGTGGTTCAGGATGAATTACAGTATTTCACCCTTGCCCTTGACCTC TACTTCAAGTATAACGTTACGGAAATGCTGTCAAGTGGATGGATACAGGTTTCAAATGGTAAGGAATATGCACTGAGCGA TGTCATCGATACCATCAAACATGCTTTTGGGGGATCACCACAAATTGTTTGCAAGAGGGGTTCAATCGAAGAACTTAGGT TATGCTTCGACAAAGAATTGAAGCCTCGTGATTGCCTTACCACTTCTTTGGCGAATGGAAGTGTATCAAAAAGCAAGCAC TGCCCGCGGTACATCACCTTGCCAACGTATGATCCCCTTGTGCTTGCCAATTCAACTGTAGAGATCATGACACAGTTCGA TGAATTTGAGGTGCCTGCAGCTCTTTATACAGCCTAA |