
| Microexon ID | Ps_NC_039362.1:211670940-211670943:- |
| Species | Papaver somniferum | Coordinates | NC_039362.1:211670940..211670943 |
| Microexon Cluster ID | MEP04 |
| Size | 4 |
| Phase | 0 |
| Pfam Domain Motif | Ribonuclease_T2 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 51,4,53 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RGYWMWYCATCAACHTGCYWYRGTGGAAARGGAYYATTYTGGGCTCATGAGTGGGARAARCATGGRACWTGYTCYTCYCCWGTWRTTCRWGATGAATAYARYTAYTTY |
| Logo of Microexon-tag DNA Seq | 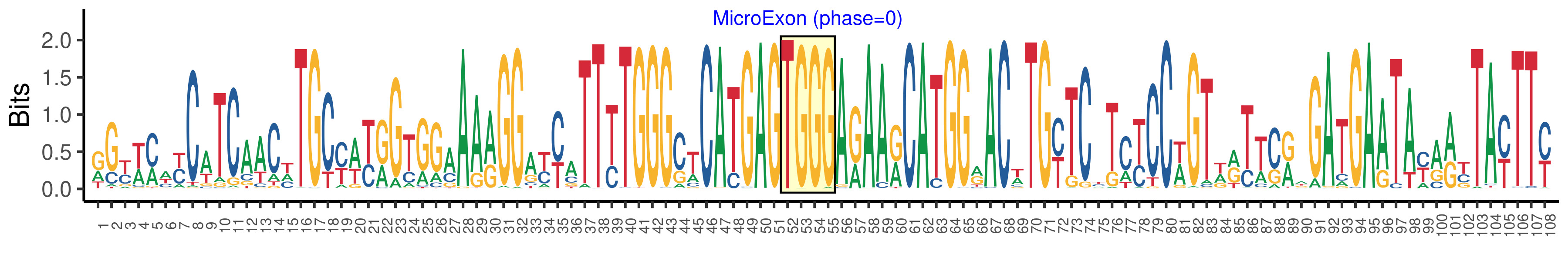 |
| Alignment of exons | 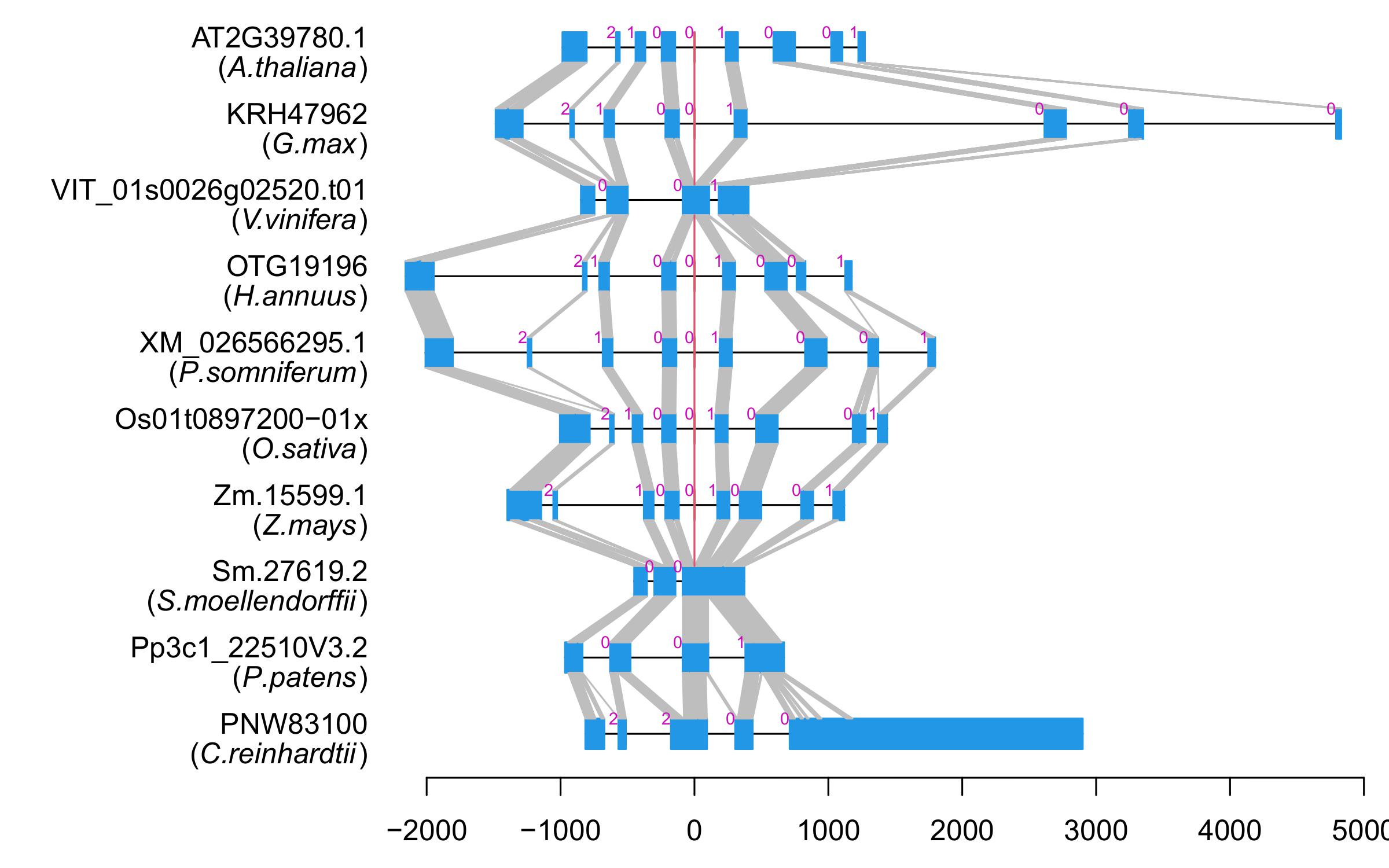 |
| Microexon DNA seq | TGGG |
| Microexon Amino Acid seq | WE |
| Microexon-tag DNA Seq | GGAAAGCCATCGACCTGCAGTGGTGGAAGAGGACTCTTCTGGGCCCATGAGTGGGAAAAGCATGGCACATGTTCTTCTCCAGTAGTTCATGATGAATACCAGTACTTC |
| Microexon-tag Amino Acid Seq | GKPSTCSGGRGLFWAHEWEKHGTCSSPVVHDEYQYF |
| Microexon-tag spanning region | 211670692-211671127 |
| Microexon-tag prediction score | 0.9613 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026534029.1x |
| Reference Transcript ID | XM_026534029.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026534029.1 |
| Protein ID | XP_026389814.1 |
| Gene ID | LOC113284533 |
| Gene Name | NA |
| Pfam domain motif | Ribonuclease_T2 |
| Motif E-value | 1.2e-49 |
| Motif start | 40 |
| Motif end | 227 |
| Protein seq | >XP_026389814.1 MAFSKTLFVSVLLIVLGSCLFVEIDGRSRSRSLLSAQREFDYFALSLQWPGTICQHTRHCCAGNGCCRSDNGPTEFTIHG LWPDYNDGTWPACCTETRFDEKLISSLRKDLDKYWPSLSCGKPSTCSGGRGLFWAHEWEKHGTCSSPVVHDEYQYFTTTL SVYFKYNVTAILAAEGYVPSNSEKYPLGGIVSAVQNAFGATPQVVCSHGAIEELRLCFYKDFKPRDCIMNEKHRSCPKYV SMPERRPMGHESALPWSLNMDT* |
| CDS seq | >XM_026534029.1 ATGGCTTTCTCTAAAACTCTTTTTGTCTCTGTTCTCTTAATTGTATTGGGTTCTTGTTTGTTCGTCGAAATTGATGGTAG AAGTAGAAGCAGAAGTTTATTGAGTGCACAAAGAGAGTTTGATTACTTTGCTTTATCTCTTCAATGGCCTGGTACTATTT GTCAACATACTAGACATTGCTGTGCTGGTAATGGTTGTTGCAGAAGTGATAATGGACCAACAGAGTTCACCATCCATGGA CTCTGGCCTGACTACAATGATGGAACCTGGCCAGCATGTTGCACCGAAACTAGATTTGATGAGAAATTGATCTCATCATT GCGCAAAGATCTAGACAAGTATTGGCCGTCCTTAAGCTGTGGAAAGCCATCGACCTGCAGTGGTGGAAGAGGACTCTTCT GGGCCCATGAGTGGGAAAAGCATGGCACATGTTCTTCTCCAGTAGTTCATGATGAATACCAGTACTTCACAACAACTCTC AGTGTTTACTTCAAGTATAACGTCACGGCAATTTTGGCTGCCGAAGGATATGTACCTTCAAATTCTGAGAAATATCCCTT GGGAGGCATTGTTTCAGCCGTTCAAAATGCTTTTGGAGCAACTCCTCAGGTGGTTTGCTCACATGGTGCCATAGAGGAGC TTCGCTTATGCTTTTACAAAGATTTCAAGCCTCGCGACTGCATAATGAATGAAAAGCATAGGTCATGCCCCAAATACGTG AGCATGCCAGAACGCAGGCCGATGGGACATGAGTCTGCACTTCCATGGTCACTTAATATGGATACATAA |
| Microexon DNA seq | TGGG |
| Microexon Amino Acid seq | WE |
| Microexon-tag DNA Seq | GGAAAGCCATCGACCTGCAGTGGTGGAAGAGGACTCTTCTGGGCCCATGAGTGGGAAAAGCATGGCACATGTTCTTCTCCAGTAGTTCATGATGAATACCAGTACTTC |
| Microexon-tag Amino Acid seq | GKPSTCSGGRGLFWAHEWEKHGTCSSPVVHDEYQYF |
| Transcript ID | XM_026534029.1 |
| Gene ID | Ps.33633 |
| Gene Name | NA |
| Pfam domain motif | Ribonuclease_T2 |
| Motif E-value | 1.2e-49 |
| Motif start | 40 |
| Motif end | 227 |
| Protein seq | >XM_026534029.1 MAFSKTLFVSVLLIVLGSCLFVEIDGRSRSRSLLSAQREFDYFALSLQWPGTICQHTRHCCAGNGCCRSDNGPTEFTIHG LWPDYNDGTWPACCTETRFDEKLISSLRKDLDKYWPSLSCGKPSTCSGGRGLFWAHEWEKHGTCSSPVVHDEYQYFTTTL SVYFKYNVTAILAAEGYVPSNSEKYPLGGIVSAVQNAFGATPQVVCSHGAIEELRLCFYKDFKPRDCIMNEKHRSCPKYV SMPERRPMGHESALPWSLNMDT* |
| CDS seq | >XM_026534029.1 ATGGCTTTCTCTAAAACTCTTTTTGTCTCTGTTCTCTTAATTGTATTGGGTTCTTGTTTGTTCGTCGAAATTGATGGTAG AAGTAGAAGCAGAAGTTTATTGAGTGCACAAAGAGAGTTTGATTACTTTGCTTTATCTCTTCAATGGCCTGGTACTATTT GTCAACATACTAGACATTGCTGTGCTGGTAATGGTTGTTGCAGAAGTGATAATGGACCAACAGAGTTCACCATCCATGGA CTCTGGCCTGACTACAATGATGGAACCTGGCCAGCATGTTGCACCGAAACTAGATTTGATGAGAAATTGATCTCATCATT GCGCAAAGATCTAGACAAGTATTGGCCGTCCTTAAGCTGTGGAAAGCCATCGACCTGCAGTGGTGGAAGAGGACTCTTCT GGGCCCATGAGTGGGAAAAGCATGGCACATGTTCTTCTCCAGTAGTTCATGATGAATACCAGTACTTCACAACAACTCTC AGTGTTTACTTCAAGTATAACGTCACGGCAATTTTGGCTGCCGAAGGATATGTACCTTCAAATTCTGAGAAATATCCCTT GGGAGGCATTGTTTCAGCCGTTCAAAATGCTTTTGGAGCAACTCCTCAGGTGGTTTGCTCACATGGTGCCATAGAGGAGC TTCGCTTATGCTTTTACAAAGATTTCAAGCCTCGCGACTGCATAATGAATGAAAAGCATAGGTCATGCCCCAAATACGTG AGCATGCCAGAACGCAGGCCGATGGGACATGAGTCTGCACTTCCATGGTCACTTAATATGGATACATAA |